5LS0
 
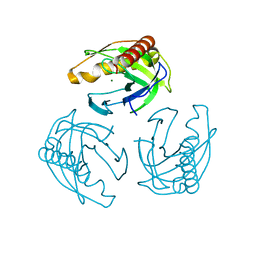 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | Authors: | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
4LUG
 
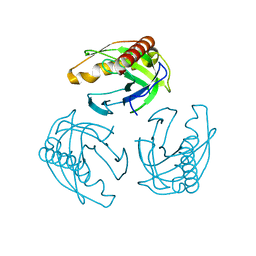 | |
7Z0U
 
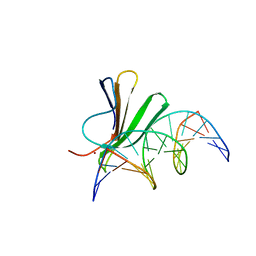 | | Crystal structure of AtWRKY18 DNA-binding domain in complex with W-box DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*GP*GP*CP*G)-3'), WRKY transcription factor 18, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
7Z0R
 
 | | AtWRKY18 DNA-binding domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NITRATE ION, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
6ZTX
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZTW
 
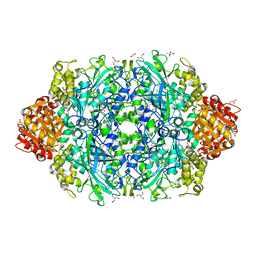 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
8OWM
 
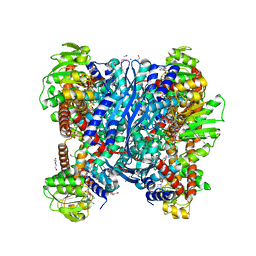 | | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(oxidanyl)pentanedioic acid, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site.
Plant Physiol Biochem., 201, 2023
|
|
7QB6
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with 3-carboxybenzophenone | | Descriptor: | 3-benzoylbenzoic acid, MALONATE ION, Nodulin-13 | | Authors: | Grzechowiak, M, Ignasiak, M, Nowicka-Bauer, K, Marciniak, B, Jaskolski, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Does the presence of ground state complex between a PR-10 protein and a sensitizer affect the mechanism of sensitized photo-oxidation?
Free Radic Biol Med, 198, 2023
|
|
8OWN
 
 | |
8S3D
 
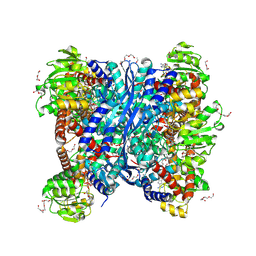 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with 2-amino-2-hydroxyglutarate (reaction intermediate) and NAD | | Descriptor: | (2S)-2-azanyl-2-oxidanyl-pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S3A
 
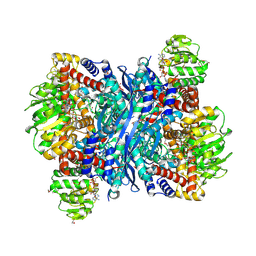 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with 2,6-pyridinedicarboxylic acid and NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S3B
 
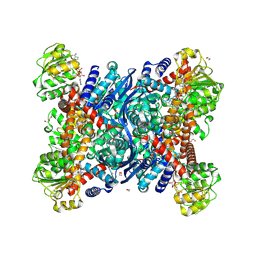 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with 3-(1H-Tetrazol-5-yl)benzoic acid and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(1~{H}-1,2,3,4-tetrazol-5-yl)benzoic acid, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S38
 
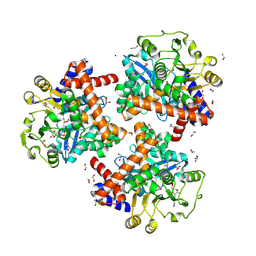 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with citrate and NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S39
 
 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with isophthalic acid and NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S3C
 
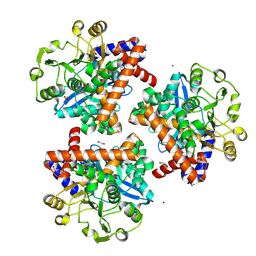 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 (unliganded) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
9G67
 
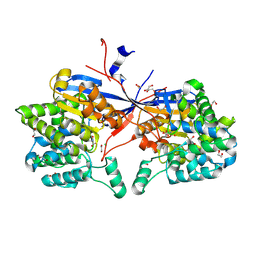 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138H mutant in complex with L-Asn | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CADMIUM ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-07-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling enzyme activity by mutagenesis and metal exchange to obtain crystal structures of stable substrate complexes of Class 3 l-asparaginase.
Febs J., 292, 2025
|
|
9G68
 
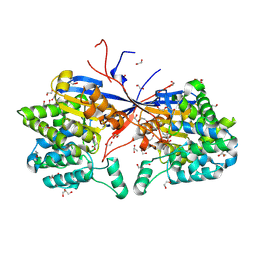 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138A mutant in complex with L-Asn | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-07-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Controlling enzyme activity by mutagenesis and metal exchange to obtain crystal structures of stable substrate complexes of Class 3 l-asparaginase.
Febs J., 292, 2025
|
|
9G66
 
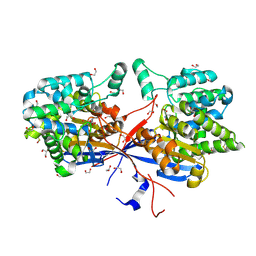 | | Crystal structure of WT Rhizobium etli L-asparaginase ReAV in complex with L-Asn | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CADMIUM ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-07-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Controlling enzyme activity by mutagenesis and metal exchange to obtain crystal structures of stable substrate complexes of Class 3 l-asparaginase.
Febs J., 292, 2025
|
|
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
6YEI
 
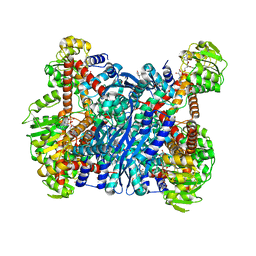 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
6MT1
 
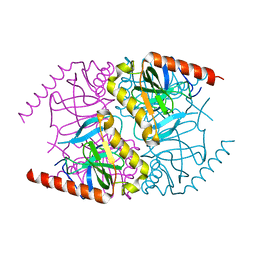 | |
6MT2
 
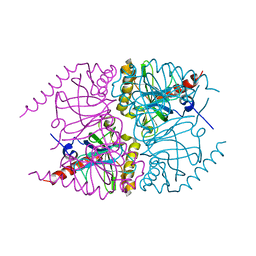 | |
8CLZ
 
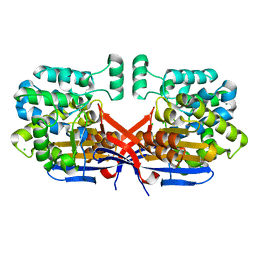 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (monoclinic form R4mC-2) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8COL
 
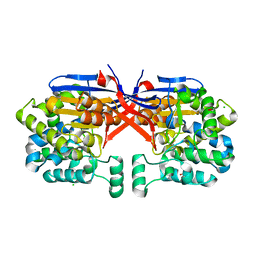 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (orthorombic form R4oP-2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imioloczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
