4GK7
 
 | | yeast 20S proteasome in complex with the Syringolin-Glidobactin chimera | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Stein, M.L, Bachmann, A. | | Deposit date: | 2012-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity enhancement of the synthetic syrbactin proteasome inhibitor hybrid and biological evaluation in tumor cells.
Biochemistry, 51, 2012
|
|
1RYP
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
1G0U
 
 | | A GATED CHANNEL INTO THE PROTEASOME CORE PARTICLE | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Bajorek, M, Kohler, A, Moroder, L, Rubin, D.M, Huber, R, Glickman, M.H, Finley, D. | | Deposit date: | 2000-10-09 | | Release date: | 2000-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gated channel into the proteasome core particle.
Nat.Struct.Biol., 7, 2000
|
|
1G65
 
 | | Crystal structure of epoxomicin:20s proteasome reveals a molecular basis for selectivity of alpha,beta-epoxyketone proteasome inhibitors | | Descriptor: | EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, Proteasome component C1, ... | | Authors: | Groll, M, Kim, K.B, Kairies, N, Huber, R, Crews, C. | | Deposit date: | 2000-11-03 | | Release date: | 2000-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Epoxomicin:20S Proteasome reveals a molecular basis for selectivity of alpha,beta-Epoxyketone Proteasome Inhibitors
J.Am.Chem.Soc., 122, 2000
|
|
2GPL
 
 | | TMC-95 based biphenyl-ether macrocycles: specific proteasome inhibitors | | Descriptor: | BENZYL [12-(2-AMINO-2-OXOETHYL)-4-NITRO-10,13-DIOXO-15-[(PROPYLAMINO)CARBONYL]-2-OXA-11,14-DIAZATRICYCLO[15 .2.2.1~3,7~]DOCOSA-1(19),3(22),4,6,17,20-HEXAEN-9-YL]CARBAMATE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Goetz, M, Kaiser, M, Weyher, E, Moroder, M. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TMC-95-Based Inhibitor Design Provides Evidence for the Catalytic Versatility of the Proteasome.
Chem.Biol., 13, 2006
|
|
2R2Y
 
 | |
7NCB
 
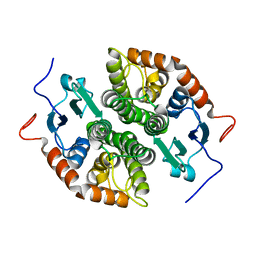 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC9
 
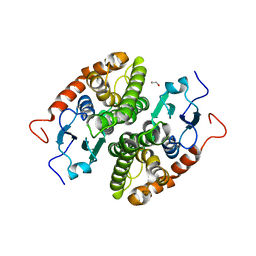 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCM
 
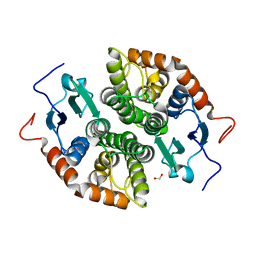 | | Glutathione-S-transferase GliG mutant E82A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCU
 
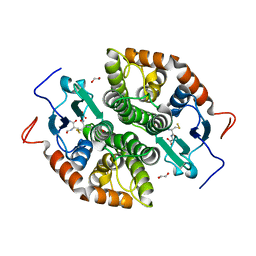 | |
7NCT
 
 | | Glutathione-S-transferase GliG mutant K127G | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC2
 
 | |
7NCE
 
 | | Glutathione-S-transferase GliG mutant N27A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCL
 
 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC1
 
 | |
7NCD
 
 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC8
 
 | | Glutathione-S-transferase GliG mutant S24A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC6
 
 | | Glutathione-S-transferase GliG in complex with cyclo[L-Phe-L-Ser]-bis-glutathione-adduct | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-3-[(2~{R},5~{R})-5-(hydroxymethyl)-3,6-bis(oxidanylidene)-2-(phenylmethyl)-5-sulfanyl-piperazin-2-yl]sulfanyl-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCO
 
 | | Glutathione-S-transferase GliG mutant K127R | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC5
 
 | |
7NCN
 
 | | Glutathione-S-transferase GliG mutant S83A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC3
 
 | |
7NCP
 
 | | Glutathione-S-transferase GliG mutant K127A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG, SODIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2ZCY
 
 | | yeast 20S proteasome:syringolin A-complex | | Descriptor: | (2S)-2-[[(2S)-1-[[(5S,8S,9E)-2,7-dioxo-5-propan-2-yl-1,6-diazacyclododeca-3,9-dien-8-yl]amino]-3-methyl-1-oxo-butan-2-yl]carbamoylamino]-3-methyl-butanoic acid, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
3BDM
 
 | | yeast 20S proteasome:glidobactin A-complex | | Descriptor: | (2E,4E)-N-[(2S,3R)-3-hydroxy-1-[[(3Z,5S,8S,10S)-10-hydroxy-5-methyl-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]amino]-1-ox obutan-2-yl]dodeca-2,4-dienamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
