2J7O
 
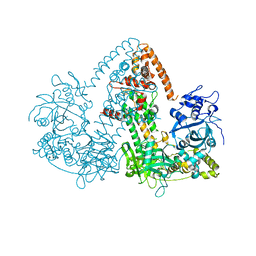 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2J7N
 
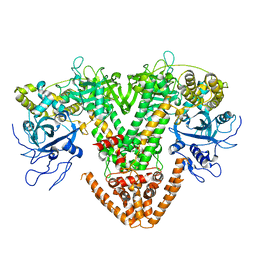 | | Structure of the RNAi polymerase from Neurospora crassa | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2JHP
 
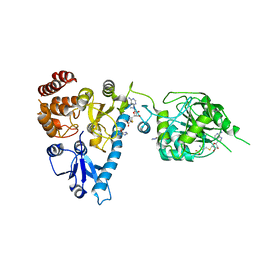 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JH9
 
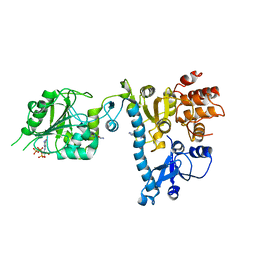 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, GUANOSINE-5'-TRIPHOSPHATE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JH8
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7NHX
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIS
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core | | Descriptor: | Nanobody8192 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK1
 
 | | 1918 Influenza virus polymerase heterotirmer in complex with vRNA promoters and Nb8201 | | Descriptor: | Nanobody8201, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NFR
 
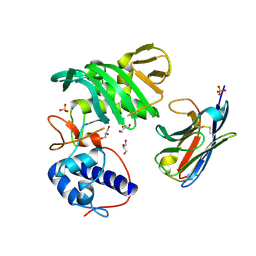 | | Fujian capmidlink domain in complex with Nb8194 | | Descriptor: | GLYCEROL, NB8194, Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKI
 
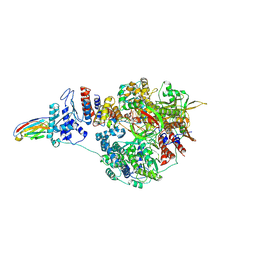 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8209 core | | Descriptor: | Nb8209, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK6
 
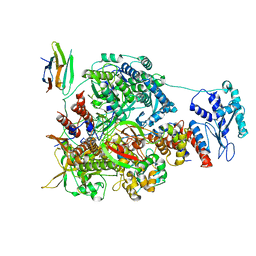 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8204 | | Descriptor: | Nb8204, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (6.72 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKR
 
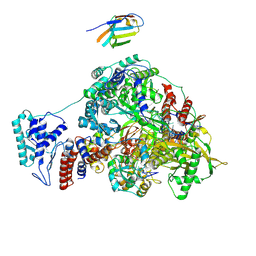 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8210 | | Descriptor: | Nb8210, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ7
 
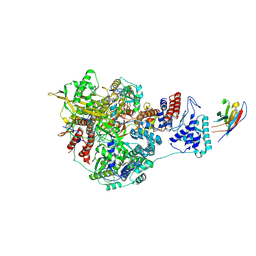 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8200 core | | Descriptor: | Nb8200 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.82 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8206 | | Descriptor: | NB8206, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NHA
 
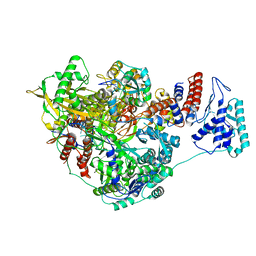 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Endonuclease and priming loop ordered (Class2a) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIK
 
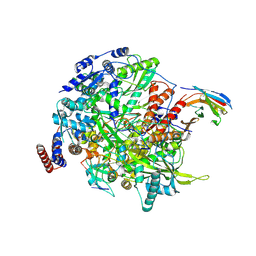 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8189 core | | Descriptor: | Nanobody8189 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ5
 
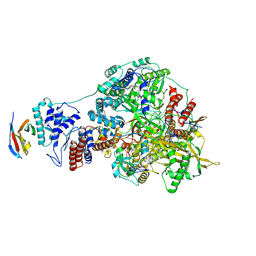 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8199 core | | Descriptor: | Nb8199 Core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKC
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8207 | | Descriptor: | Nb8207, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NI0
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Replicase (class 3) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8191 core | | Descriptor: | Nanobody8191 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NHC
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Endonuclease ordered (Class2b) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIL
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8190 core | | Descriptor: | Nanobody8190 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK2
 
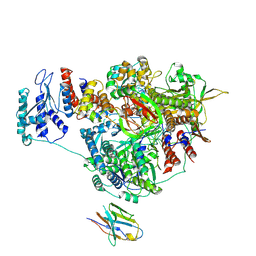 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8202 core | | Descriptor: | Nanobody8202, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ3
 
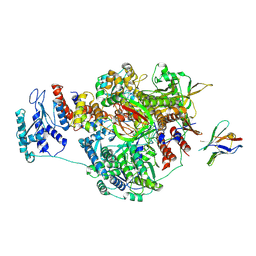 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8196 core | | Descriptor: | Nanobody8196 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-15 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ4
 
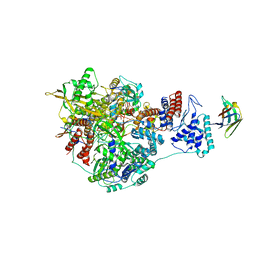 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8198 core | | Descriptor: | Nb8198 Core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
