4A6A
 
 | |
3PNI
 
 | |
7LZI
 
 | |
7LZ2
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZH
 
 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
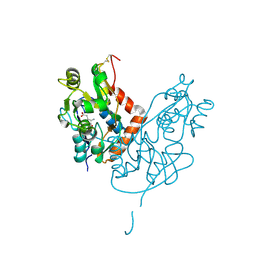 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ1
 
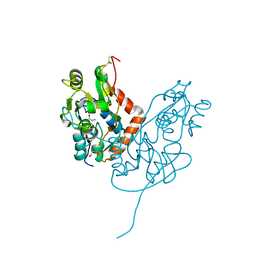 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
6VEA
 
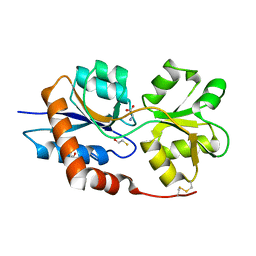 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VE8
 
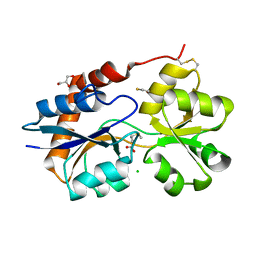 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
