5V6O
 
 | |
5V6N
 
 | |
3UQ4
 
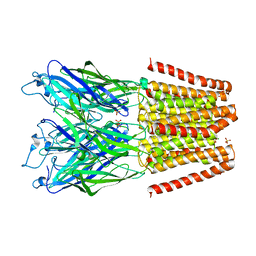
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant F247L (F16L) | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQ7
 
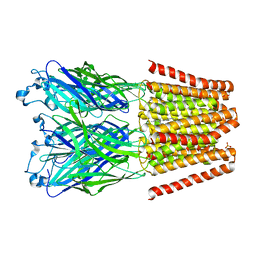
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240S F247L (L9S F16L) in presence of 10 mM cysteamine | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQ5
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240A F247L (L9A F16L) in the presence of 10 mM cysteamine | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7TE2
 
 | | Crystal Structure of AerR from Rhodobacter capsulatus at 2.25 A. | | Descriptor: | AerR, CHLORIDE ION, COBALAMIN, ... | | Authors: | Dragnea, V, Gonzalez-Gutierrez, G, Bauer, C.E. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analyses of CrtJ and Its B 12 -Binding Co-Regulators SAerR and LAerR from the Purple Photosynthetic Bacterium Rhodobacter capsulatus.
Microorganisms, 10, 2022
|
|
6CDB
 
 | | Crystal Structure of V66L CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, SODIUM ION, ... | | Authors: | Capdevila, D.A, Campanello, G, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
6PHN
 
 | |
6PHI
 
 | |
6PHM
 
 | |
6PHL
 
 | |
6PHQ
 
 | |
6MNZ
 
 | | Crystal structure of RibBX, a two domain 3,4-dihydroxy-2-butanone 4-phosphate synthase from A. baumannii. | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Wang, J, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multi-metal Restriction by Calprotectin Impacts De Novo Flavin Biosynthesis in Acinetobacter baumannii.
Cell Chem Biol, 26, 2019
|
|
6WAI
 
 | | Crystal Structure of SmcR N142D from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAE
 
 | | Crystal Structure of 6X-His tagged SmcR | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAG
 
 | | Crystal Structure of SmcR S76A from Vibrio Vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAF
 
 | | Crystal Structure of SmcR N55I from Vibrio vulnificus | | Descriptor: | LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.381 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
6WAH
 
 | | Crystal Structure of SmcR L139R from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
7MQ2
 
 | |
7MQ3
 
 | |
7MQ1
 
 | | C9A Streptococcus pneumoniae CstR in the reduced state, space group C2 | | Descriptor: | CHLORIDE ION, Copper-sensing transcriptional repressor csoR, GLYCEROL, ... | | Authors: | Fakhoury, J.N, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional asymmetry and chemical reactivity of CsoR family persulfide sensors.
Nucleic Acids Res., 49, 2021
|
|
6E5G
 
 | |
4LMK
 
 | |
4LMJ
 
 | |
4LML
 
 | | GLIC double mutant I9'A T25'A | | Descriptor: | Proton-gated ion channel | | Authors: | Grosman, C, Gonzalez-Gutierrez, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
