3QY9
 
 | |
2PI7
 
 | |
4EWT
 
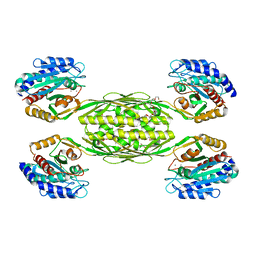 | | The crystal structure of a putative aminohydrolase from methicillin resistant Staphylococcus aureus | | Descriptor: | 1-DEOXY-1-THIO-HEPTAETHYLENE GLYCOL, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Girish, T.S, Vivek, B, Colaco, M, Misquith, S, Gopal, B. | | Deposit date: | 2012-04-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an amidohydrolase, SACOL0085, from methicillin-resistant Staphylococcus aureus COL
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3KI9
 
 | |
3KHX
 
 | |
3KHZ
 
 | |
