7GT4
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000528a | | Descriptor: | (4R)-4-hydroxy-2-(2-hydroxyethyl)-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTG
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000684a | | Descriptor: | (5R,7S,8R,8aS)-2-(cyclopropylmethyl)-8-phenyloctahydropyrrolo[1,2-a]pyrazine-7-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GU9
 
 | |
7GST
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000056a | | Descriptor: | 1-(methanesulfonyl)-1,2,3,4-tetrahydroquinoline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTN
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000625a | | Descriptor: | 1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTZ
 
 | |
7GU6
 
 | |
7GSC
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000729a | | Descriptor: | (azepan-1-yl)(2,6-difluorophenyl)methanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSN
 
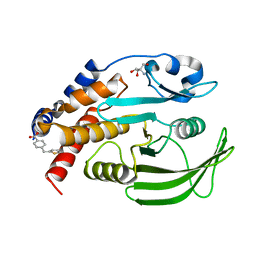 | |
7GT2
 
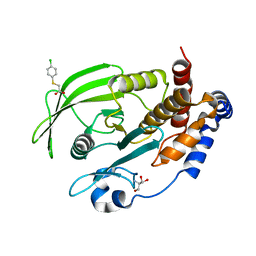 | |
7GU1
 
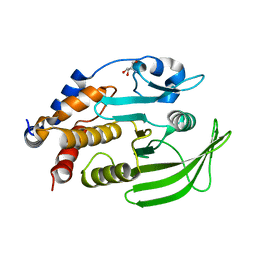 | |
7GUB
 
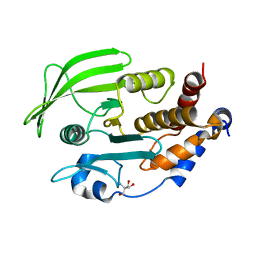 | |
7GUA
 
 | |
7GTB
 
 | |
7GTJ
 
 | |
7GUC
 
 | |
7GSV
 
 | |
7GT1
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000209a | | Descriptor: | (2S)-2-(2-chloro-6-fluorophenyl)-2,3-dihydroquinazolin-4(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT6
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000530a | | Descriptor: | (3aS,8aS)-6-benzoyloctahydropyrrolo[3,4-d]azepin-1(2H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTE
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000646b | | Descriptor: | (5S)-5-(trifluoromethyl)-1,4-diazepane, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT3
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000527a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ethyl (3R,3aS,8bS)-1-acetyl-5-methyl-2,3,3a,8b-tetrahydro-1H-[1]benzofuro[3,2-b]pyrrole-3-carboxylate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT9
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000463b | | Descriptor: | (3R)-4-oxo-3,4-dihydro-2H-1-benzopyran-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTM
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000543a | | Descriptor: | (4S)-4-hydroxy-2-(propan-2-yl)-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSI
 
 | |
7GTI
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000571a | | Descriptor: | 1-{(1S,4R,5S,6R)-6-hydroxy-4-[(pyridin-2-yl)oxy]-2-azabicyclo[3.3.1]nonan-2-yl}ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
