3G5L
 
 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
3G2E
 
 | | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from Campylobacter jejuni | | Descriptor: | GLYCEROL, OORC subunit of 2-oxoglutarate:acceptor oxidoreductase | | Authors: | Ramagopal, U.A, Toro, R, Miller, S, Gilmore, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from
Campylobacter jejuni
To be Published
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | Descriptor: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
3CE2
 
 | | Crystal structure of putative peptidase from Chlamydophila abortus | | Descriptor: | Putative peptidase, ZINC ION | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Eberle, M, Maletic, M, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative peptidase from Chlamydophila abortus.
To be Published
|
|
3C9F
 
 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | Descriptor: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
3ED4
 
 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
3EQZ
 
 | | Crystal structure of a response regulator from Colwellia psychrerythraea | | Descriptor: | Response regulator | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a response regulator from Colwellia psychrerythraea
To be Published
|
|
3EMU
 
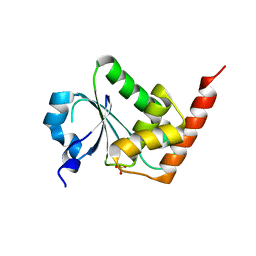 | | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica | | Descriptor: | SULFATE ION, leucine rich repeat and phosphatase domain containing protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica
To be Published
|
|
3CAX
 
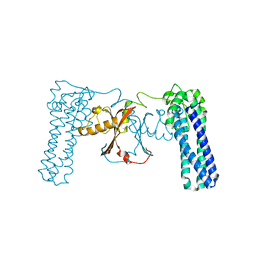 | | Crystal structure of uncharacterized protein PF0695 | | Descriptor: | Uncharacterized protein PF0695 | | Authors: | Ramagopal, U.A, Hu, S, Toro, R, Gilmore, M, Bain, K, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of uncharacterized protein PF0695.
To be Published
|
|
3EGO
 
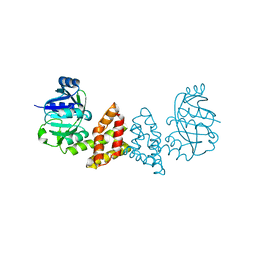 | | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis | | Descriptor: | Probable 2-dehydropantoate 2-reductase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-11 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis
To be published
|
|
3EGC
 
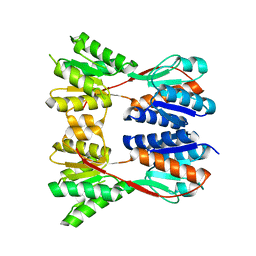 | | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis | | Descriptor: | putative ribose operon repressor | | Authors: | Bonanno, J.B, Patskovsky, Y, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis
To be Published
|
|
3EWB
 
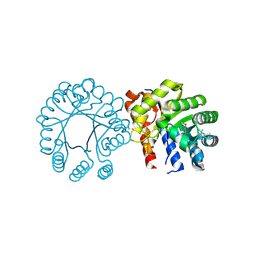 | | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes | | Descriptor: | 2-isopropylmalate synthase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes.
To be Published
|
|
3CG0
 
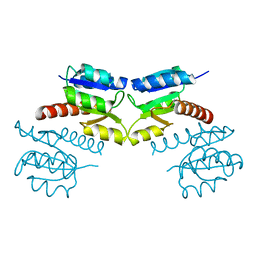 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3EVY
 
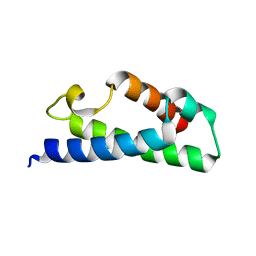 | | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis | | Descriptor: | Putative type I restriction enzyme R protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis
To be Published
|
|
3F1C
 
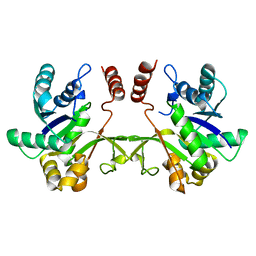 | | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes | | Descriptor: | Putative 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase 2 | | Authors: | Patskovsky, Y, Ho, J, Toro, R, Gilmore, M, Miller, S, Groshong, C, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes
To be Published
|
|
3FHL
 
 | | Crystal structure of a putative oxidoreductase from bacteroides fragilis nctc 9343 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative oxidoreductase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-09 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase from Bacteroides Fragilis Nctc 9343
To be Published
|
|
3EVT
 
 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
3FBG
 
 | | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus | | Descriptor: | MAGNESIUM ION, putative arginate lyase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus
To be Published
|
|
3FNR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | Descriptor: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-26 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
3EEQ
 
 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
3FRN
 
 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
3GFG
 
 | | Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form | | Descriptor: | Uncharacterized oxidoreductase yvaA | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Chang, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form.
To be published
|
|
