1AJ9
 
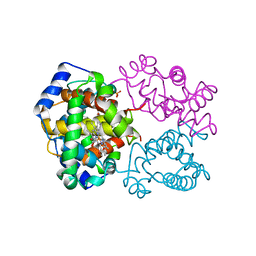 | | R-STATE HUMAN CARBONMONOXYHEMOGLOBIN ALPHA-A53S | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human carboxyhemoglobin at 2.2 A resolution: structure and solvent comparisons of R-state, R2-state and T-state hemoglobins.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3NCJ
 
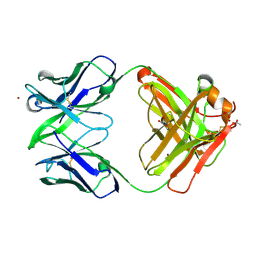 | | Crystal structure of Fab15 Mut8 | | Descriptor: | ACETATE ION, Fab15 Mut8 heavy chain, Fab15 Mut8 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NAA
 
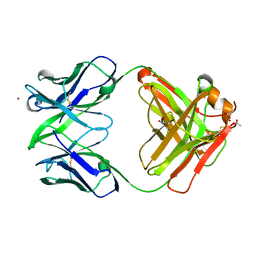 | | Crystal structure of Fab15 Mut5 | | Descriptor: | ACETATE ION, Fab15 Mut5 heavy chain, Fab15 Mut5 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3O11
 
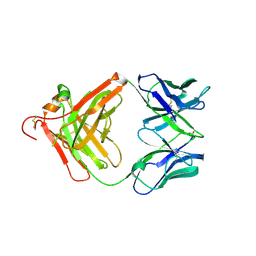 | | Anti-beta-amyloid antibody c706 fab in space group c2 | | Descriptor: | C706 HEAVY CHAIN variable region, Ig gamma-1 chain C region chimera, C706 LIGHT CHAIN variable region, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | His-tag binding by antibody C706 mimics beta-amyloid recognition.
J.Mol.Recognit., 24, 2011
|
|
31BI
 
 | |
1HW5
 
 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
1HDB
 
 | | ANALYSIS OF THE CRYSTAL STRUCTURE, MOLECULAR MODELING AND INFRARED SPECTROSCOPY OF THE DISTAL BETA-HEME POCKET VALINE67(E11)-THREONINE MUTATION OF HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) BETA-V67T, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pechik, I, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1995-04-14 | | Release date: | 1996-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic, molecular modeling, and biophysical characterization of the valine beta 67 (E11)-->threonine variant of hemoglobin.
Biochemistry, 35, 1996
|
|
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1R4B
 
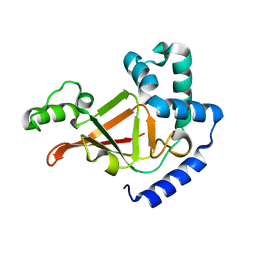 | |
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
1PGA
 
 | |
1S4C
 
 | | YHCH PROTEIN (HI0227) COPPER COMPLEX | | Descriptor: | ACETATE ION, COPPER (II) ION, Protein HI0227 | | Authors: | Teplyakov, A, Obmolova, G, Toedt, J, Gilliland, G.L. | | Deposit date: | 2004-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
J.Bacteriol., 187, 2005
|
|
1R4W
 
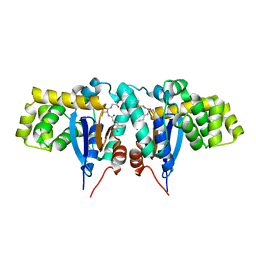 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1PGB
 
 | |
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1SUA
 
 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1SUE
 
 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
1CLS
 
 | | CROSS-LINKED HUMAN HEMOGLOBIN DEOXY | | Descriptor: | HEMOGLOBIN, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ji, X, Fronticelli, C, Bucci, E, Gilliland, G.L. | | Deposit date: | 1995-08-29 | | Release date: | 1996-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive and negative cooperativities at subsequent steps of oxygenation regulate the allosteric behavior of multistate sebacylhemoglobin.
Biochemistry, 35, 1996
|
|
1N2A
 
 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1MTC
 
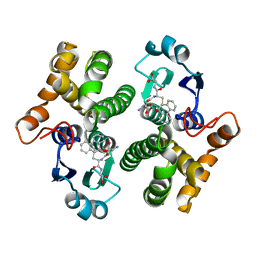 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
1SUP
 
 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1RW1
 
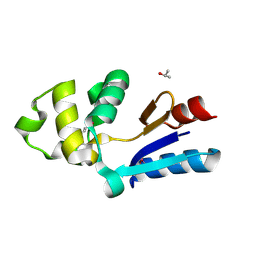 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1NRK
 
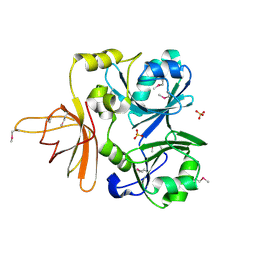 | |
1SUC
 
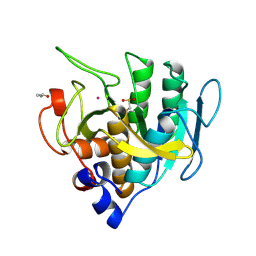 | |
