5GIQ
 
 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5GIU
 
 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | Authors: | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
5XEV
 
 | | Crystal Structure of a novel Xaa-Pro dipeptidase from Deinococcus radiodurans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Are, V.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a novel prolidase from Deinococcus radiodurans identifies new subfamily of bacterial prolidases.
Proteins, 85, 2017
|
|
5GIV
 
 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
5X49
 
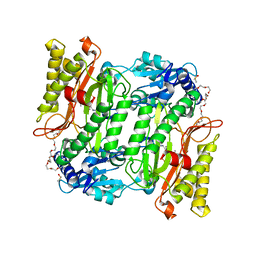 | | Crystal Structure of Human mitochondrial X-prolyl Aminopeptidase (XPNPEP3) | | Descriptor: | (2S,3R)-3-amino-2-hydroxy-4-phenylbutanoic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Singh, R, Kumar, A, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the human aminopeptidase XPNPEP3 and comparison of its in vitro activity with Icp55 orthologs: Insights into diverse cellular processes.
J. Biol. Chem., 292, 2017
|
|
7FCR
 
 | |
7FCS
 
 | |
5GGX
 
 | |
5YSC
 
 | |
4R60
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Ghosh, B, Are, V.N, Jamdar, S.N, Makde, R.D, Sharma, S.M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris
to be published
|
|
5CE6
 
 | | N-terminal domain of FACT complex subunit SPT16 from Cicer arietinum (chickpea) | | Descriptor: | ACETATE ION, FACT-Spt16, POTASSIUM ION, ... | | Authors: | Are, V.N, Ghosh, B, Kumar, A, Makde, R. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum.
Int.J.Biol.Macromol., 88, 2016
|
|
5CDL
 
 | | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-04 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative)
To Be Published
|
|
6A8M
 
 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
5FCF
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Mn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases.
Biochim. Biophys. Acta, 1865, 2017
|
|
5FCH
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
5CDV
 
 | | Proline dipeptidase from Deinococcus radiodurans R1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans R1 at 1.45 Angstrom resolution
To Be Published
|
|
5CIK
 
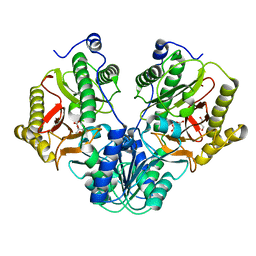 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition | | Descriptor: | CITRIC ACID, GLYCEROL, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Agrawal, U, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition
To Be Published
|
|
5CNX
 
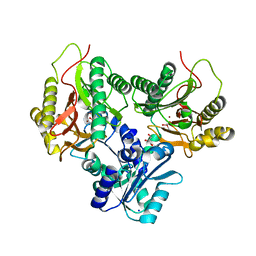 | | Crystal structure of Xaa-Pro aminopeptidase from Escherichia coli K12 | | Descriptor: | Aminopeptidase YpdF, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases
Proteins, 2018
|
|
5KHL
 
 | |
5CDE
 
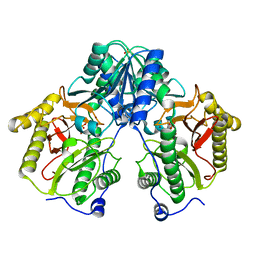 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
5GGY
 
 | |
5YZN
 
 | | Crystal structure of S9 peptidase (active form) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZM
 
 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZO
 
 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
