3JZU
 
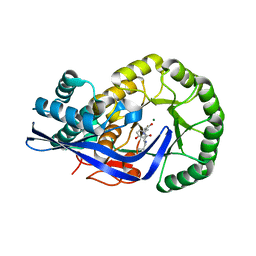 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3BOX
 
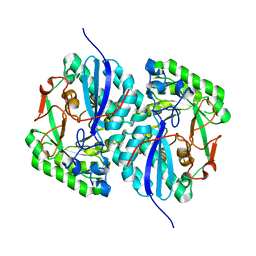 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg | | Descriptor: | L-rhamnonate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
1Q6L
 
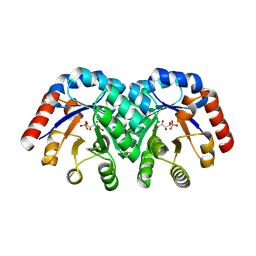 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
2PP0
 
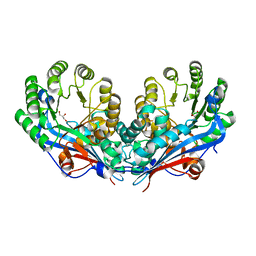 | | Crystal structure of L-talarate/galactarate dehydratase from Salmonella typhimurium LT2 | | Descriptor: | GLYCEROL, L-talarate/Galactarate Dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-04-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-talarate/galactarate dehydratase from Salmonella typhimurium LT2.
Biochemistry, 46, 2007
|
|
5T57
 
 | | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Semialdehyde dehydrogenase NAD-binding protein, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD
To be published
|
|
1BQG
 
 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
4MEV
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4MNC
 
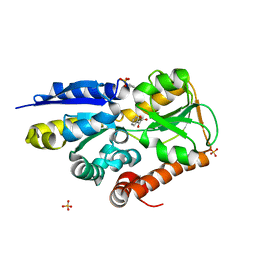 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4MNI
 
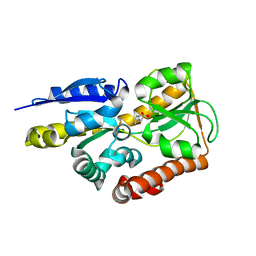 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | Descriptor: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MP4
 
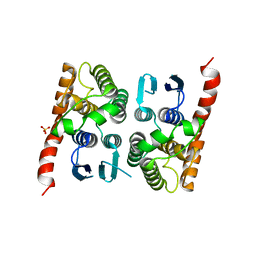 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4MK3
 
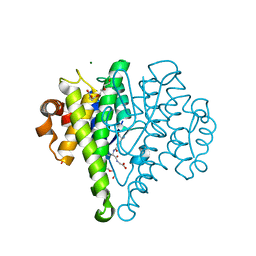 | | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h) | | Descriptor: | Glutathione S-transferase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, MAGNESIUM ION, ... | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
To be published
|
|
1JCT
 
 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JDF
 
 | | Glucarate Dehydratase from E.coli N341D mutant | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
2FLI
 
 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | Descriptor: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
2DW7
 
 | | Crystal structure of D-tartrate dehydratase from Bradyrhizobium japonicum complexed with Mg++ and meso-tartrate | | Descriptor: | Bll6730 protein, MAGNESIUM ION, S,R MESO-TARTARIC ACID | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
4MMW
 
 | | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, CHLORIDE ION, Isomerase/lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-09-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate
To be Published
|
|
2DW6
 
 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
1BFD
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
1MDR
 
 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4N2Y
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus
To be Published
|
|
