5U6G
 
 | |
6C7Z
 
 | | Crystal structure of the Q108K:K40L:T51V:R58F mutant of human Cellular Retinol Binding Protein II in complex with synthetic Ligand Julolidine | | Descriptor: | (2E,4E)-3-methyl-5-(2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)penta-2,4-dienal, ACETATE ION, Retinol-binding protein 2 | | Authors: | Nosrati, M, Geiger, J.H. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Genetically Encoded Ratiometric pH Probe: Wavelength Regulation-Inspired Design of pH Indicators.
Chembiochem, 19, 2018
|
|
1JKI
 
 | | myo-Inositol-1-phosphate Synthase Complexed with an Inhibitor, 2-deoxy-glucitol-6-phosphate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-DEOXY-GLUCITOL-6-PHOSPHATE, AMMONIUM ION, ... | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
8SDB
 
 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
4I9R
 
 | |
4LPC
 
 | |
4LQ1
 
 | |
4M6S
 
 | |
4M7M
 
 | |
4QZT
 
 | |
4QGW
 
 | | Crystal sturcture of the R132K:R111L:L121D mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.77 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
4QGV
 
 | | Crystal structure of the R132K:R111L mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.73 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
4QGX
 
 | | Crystal structure of the R132K:R111L:L121E mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.47 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
4QZU
 
 | |
4RLQ
 
 | | Crystal structure of a benzoate coenzyme A ligase with o-Toluic acid | | Descriptor: | 2-methylbenzoic acid, Benzoate-coenzyme A ligase, GLYCEROL | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K.D, Geiger, J.H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
8VZZ
 
 | |
8W02
 
 | |
8W00
 
 | |
8VZY
 
 | |
8VZX
 
 | |
2DPQ
 
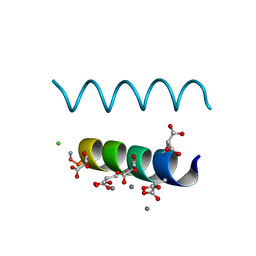 | | The crystal structures of the calcium-bound con-G and con-T(K7gamma) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, CHLORIDE ION, Conantokin-G | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
2DOI
 
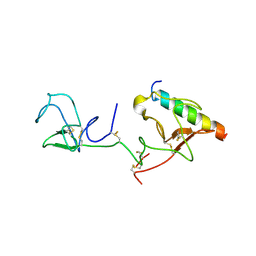 | | The X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcus protein PAM | | Descriptor: | Angiostatin, Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Cnudde, S.E, Prorok, M, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Biochemistry, 45, 2006
|
|
2FRS
 
 | |
2FS6
 
 | |
2FR3
 
 | |
