5UVJ
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
5UVK
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
5UVL
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
5UVI
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
3I17
 
 | |
4RM3
 
 | | Crystal structure of a benzoate coenzyme A ligase with 2-Furoic acid | | Descriptor: | 2-FUROIC ACID, Benzoate-coenzyme A ligase | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K.D, Geiger, J.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
4RUU
 
 | |
4RM2
 
 | | Crystal structure of a benzoate coenzyme A ligase with 2-Fluoro benzoic acid | | Descriptor: | 2-fluorobenzoic acid, Benzoate-coenzyme A ligase | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K.D, Geiger, J.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
8DN1
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V at pH 7.2 | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Borhan, B, Geiger, J.H. | | Deposit date: | 2022-07-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
2DPQ
 
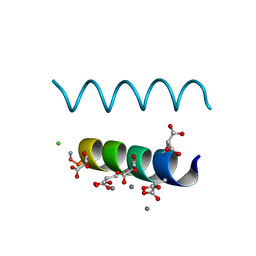 | | The crystal structures of the calcium-bound con-G and con-T(K7gamma) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, CHLORIDE ION, Conantokin-G | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
2DPR
 
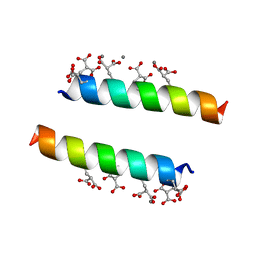 | | The crystal structures of the calcium-bound con-G and con-T(K7Glu) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, Conantokin-T | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
4GKC
 
 | |
7ML5
 
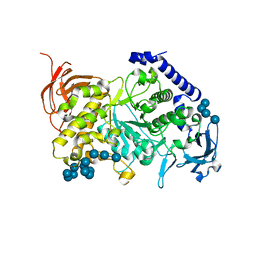 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
2DOI
 
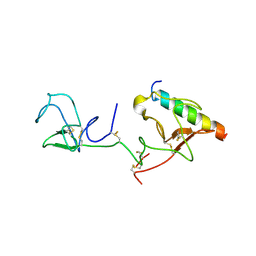 | | The X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcus protein PAM | | Descriptor: | Angiostatin, Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Cnudde, S.E, Prorok, M, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Biochemistry, 45, 2006
|
|
2DOH
 
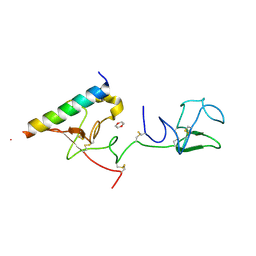 | | The X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound a to a peptide from the group A streptococcal surface protein PAM | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Angiostatin, Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Cnudde, S.E, Prorok, M, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Biochemistry, 45, 2006
|
|
7UCN
 
 | |
7UCS
 
 | |
7UCT
 
 | |
7UCV
 
 | |
7UCZ
 
 | |
7UD1
 
 | |
7UD3
 
 | |
7MS9
 
 | |
3NZ4
 
 | | Crystal Structure of a Taxus Phenylalanine Aminomutase | | Descriptor: | PHENYLETHYLENECARBOXYLIC ACID, Phenylalanine ammonia-lyase | | Authors: | Feng, L, Geiger, J.H. | | Deposit date: | 2010-07-16 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic, mutational, and structural evaluation of a taxus phenylalanine aminomutase.
Biochemistry, 50, 2011
|
|
1M7X
 
 | | The X-ray Crystallographic Structure of Branching Enzyme | | Descriptor: | 1,4-alpha-glucan Branching Enzyme | | Authors: | Abad, M.C, Binderup, K, Rios-Steiner, J, Arni, R.K, Preiss, J, Geiger, J.H. | | Deposit date: | 2002-07-23 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystallographic structure of Escherichia coli branching enzyme
J.Biol.Chem., 277, 2002
|
|
