6Q3V
 
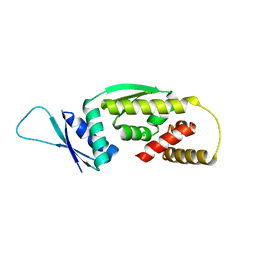 | |
6O19
 
 | | Crystal Structure of Pho7 complex with pho1 promoter site 2 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*AP*TP*TP*TP*CP*CP*GP*AP*AP*TP*AP*AP*T)-3'), DNA (5'-D(*TP*TP*AP*TP*TP*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*AP*AP*CP*A)-3'), Transcription factor Pho7, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structure of Fission Yeast Transcription Factor Pho7 Bound topho1Promoter DNA and Effect of Pho7 Mutations on DNA Binding and Phosphate Homeostasis.
Mol.Cell.Biol., 39, 2019
|
|
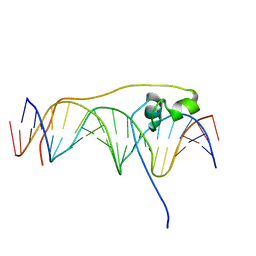
5NXT
 
 | | Wobble base paired RNA double helix | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*UP*CP*UP*AP*CP*GP*AP*AP*GP*AP*AP*CP*A)-3') | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | A novel form of RNA double helix based on G·U and C·A+ wobble base pairing.
RNA, 24, 2018
|
|
6E33
 
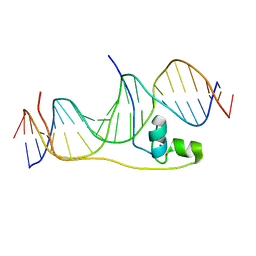 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
7NDK
 
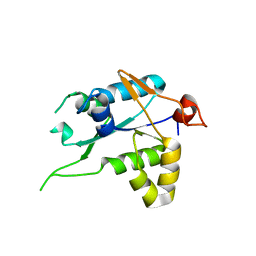 | | Crystal structure of ZC3H12C PIN catalytic mutant | | Descriptor: | Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDJ
 
 | |
7NDI
 
 | | Crystal structure of ZC3H12C PIN domain with Mg2+ Ion | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable ribonuclease ZC3H12C, ... | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDH
 
 | | Crystal structure of ZC3H12C PIN domain | | Descriptor: | 1,2-ETHANEDIOL, Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
9ASN
 
 | | Human Drosha and DGCR8 in complex with Pri-miR-98 | | Descriptor: | CALCIUM ION, Isoform 4 of Drosha, Microprocessor complex subunit DGCR8, ... | | Authors: | Garg, A, Joshua-Tor, L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs.
Mol.Cell, 84, 2024
|
|
9CXN
 
 | | Crystal structure of Human FN3K bound with AMPPNP and DMF | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(morpholin-4-yl)-D-fructose, Fructosamine-3-kinase, ... | | Authors: | Garg, A, On, K.F, Joshua-Tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9CXW
 
 | | Crystal structure of Human FN3K bound with ADP and DMF (II) | | Descriptor: | 1-deoxy-1-(morpholin-4-yl)-D-fructose, ADENOSINE-5'-DIPHOSPHATE, Fructosamine-3-kinase, ... | | Authors: | Garg, A, On, K.F, Joshua-Tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9CX8
 
 | | Crystal structure of Human FN3K in apo-state | | Descriptor: | 1,2-ETHANEDIOL, Fructosamine-3-kinase, SULFATE ION | | Authors: | Garg, A, On, K.F, Joshua-Tor, L. | | Deposit date: | 2024-07-30 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9CXM
 
 | | Crystal structure of Human FN3K bound with ATP and DMF | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(morpholin-4-yl)-D-fructose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Garg, A, On, K.F, Joshua-Tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9CXO
 
 | | Crystal structure of Human FN3K(D217S) mutant bound with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Garg, A, On, K.F, Joshua-tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9CXV
 
 | | Crystal structure of Human FN3K bound with ADP and DMF (I) | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(morpholin-4-yl)-D-fructose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Garg, A, On, K.F, Joshua-Tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
9ASP
 
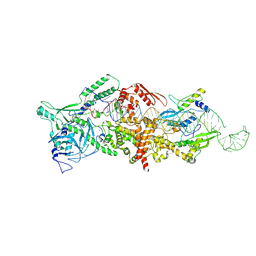 | | Human Drosha and DGCR8 in complex with Pri-let-7a1 | | Descriptor: | CALCIUM ION, Isoform 4 of Drosha, Microprocessor complex subunit DGCR8, ... | | Authors: | Garg, A, Joshua-Tor, L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs.
Mol.Cell, 84, 2024
|
|
9ASO
 
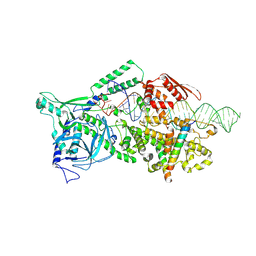 | | Human Drosha and DGCR8 in complex with Pri-let-7a2 | | Descriptor: | CALCIUM ION, Isoform 4 of Drosha, Microprocessor complex subunit DGCR8, ... | | Authors: | Garg, A, Joshua-Tor, L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs.
Mol.Cell, 84, 2024
|
|
9ASQ
 
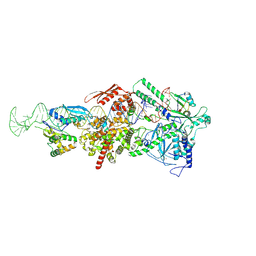 | |
9ASM
 
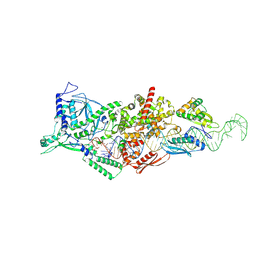 | | Human Drosha and DGCR8 in complex with Pri-let-7f1 | | Descriptor: | CALCIUM ION, Isoform 4 of Drosha, Microprocessor complex subunit DGCR8, ... | | Authors: | Garg, A, Joshua-Tor, L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs.
Mol.Cell, 84, 2024
|
|
9DUV
 
 | | Cryo-EM structure of recombinant R254H ACTA1 phalloidin-stabilized F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Garg, A, Greenberg, M.J, Zhang, R. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dilated cardiomyopathy-associated skeletal muscle actin (ACTA1) mutation R256H disrupts actin structure and function and causes cardiomyocyte hypocontractility.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DUU
 
 | | Cryo-EM structure of recombinant wildtype ACTA1 phalloidin-stabilized F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Garg, A, Greenberg, M.J, Zhang, R. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dilated cardiomyopathy-associated skeletal muscle actin (ACTA1) mutation R256H disrupts actin structure and function and causes cardiomyocyte hypocontractility.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1BTS
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTR
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1993-05-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTQ
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
