7W1I
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1J
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1L
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VPB
 
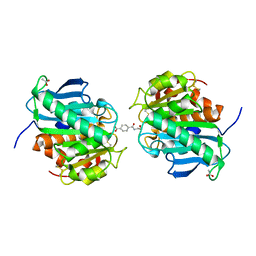 | | Crystal structure of a novel hydrolase in apo form | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, ACETATE ION, plastic degrading hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insight and engineering of a plastic degrading hydrolase Ple629.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
7X4B
 
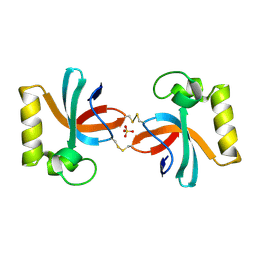 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
8WDW
 
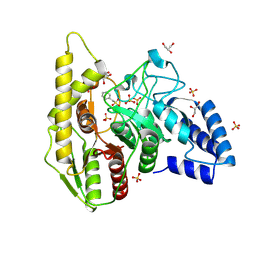 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2 | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium
To Be Published
|
|
7VME
 
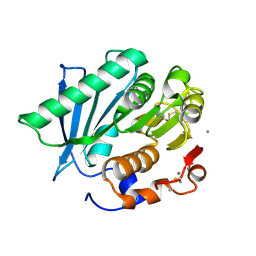 | | Crystal structure of a hydrolase in apo form 2 | | Descriptor: | CALCIUM ION, hydrolase | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of a hydrolase in apo form 2
to be published
|
|
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
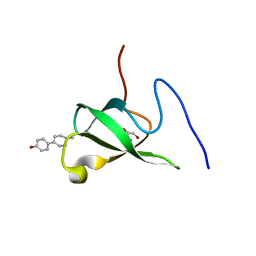 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
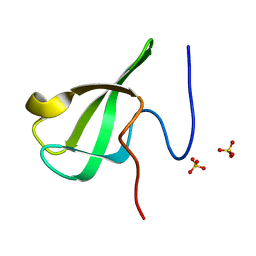 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
5XK7
 
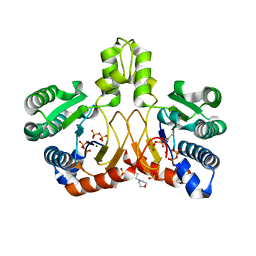 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with DMAPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK9
 
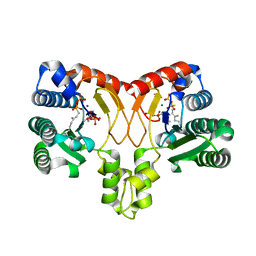 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GSPP and DMAPP | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8GW3
 
 | |
5XK8
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK6
 
 | | Structure of a prenyltransferase soaked with IPP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, SULFATE ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK3
 
 | | Crystal structure of apo form Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 | | Descriptor: | SULFATE ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6J8O
 
 | | Structure of a hypothetical protease | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structure of a hypothetical protease
To Be Published
|
|
6J8F
 
 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J9H
 
 | |
6J8N
 
 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
