3IIR
 
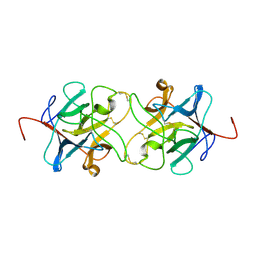 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
6HX9
 
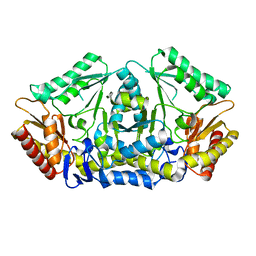 | | Putrescine transaminase from Pseudomonas putida | | Descriptor: | Aspartate aminotransferase family protein | | Authors: | Gahloth, D. | | Deposit date: | 2018-10-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of a Putrescine Transaminase FromPseudomonas putidaand its Application to the Synthesis of Benzylamine Derivatives.
Front Bioeng Biotechnol, 6, 2018
|
|
7PDA
 
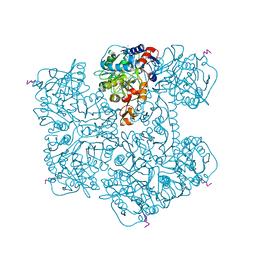 | | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-08-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum
To Be Published
|
|
7P9Q
 
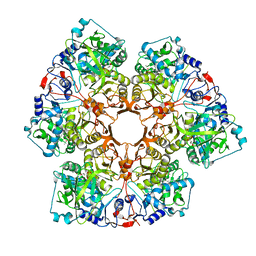 | | Crystal structure of Indole 3-Carboxylic acid decarboxylase from Arthrobacter nicotianae FI1612 in complex with co-factor prFMN. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, AnInD, MANGANESE (II) ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and biochemical characterization of the prenylated flavin mononucleotide-dependent indole-3-carboxylic acid decarboxylase.
J.Biol.Chem., 298, 2022
|
|
8PZO
 
 | | LpdD | | Descriptor: | Protein LpdD, SODIUM ION | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of LpdD from Lactobacillus plantarum.
To Be Published
|
|
8PZH
 
 | | LpdD (H61A) mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of LpdD (H61A) mutant from Lactobacillus plantarum.
To Be Published
|
|
8PO5
 
 | | Lactobacillus plantarum LpdD | | Descriptor: | MANGANESE (II) ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gallate decarboxylase subunit D, LpdD
To Be Published
|
|
8P4W
 
 | |
5MSO
 
 | |
5MSW
 
 | |
5MSP
 
 | |
5MSS
 
 | |
5MSU
 
 | |
5MST
 
 | |
5MSR
 
 | |
5MSV
 
 | |
5MJZ
 
 | |
5MK2
 
 | |
5MJY
 
 | |
5MK1
 
 | |
5MK0
 
 | |
5LOA
 
 | |
5LOC
 
 | |
