1ILY
 
 | | Solution Structure of Ribosomal Protein L18 of Thermus thermophilus | | 分子名称: | RIBOSOMAL PROTEIN L18 | | 著者 | Woestenenk, E.A, Gongadze, G.M, Shcherbakov, D.V, Rak, A.V, Garber, M.B, Hard, T, Berglund, H. | | 登録日 | 2001-05-09 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold.
Biochem.J., 363, 2002
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | 分子名称: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | 著者 | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | 登録日 | 2001-04-12 | | 公開日 | 2001-05-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF9
 
 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | 分子名称: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | 著者 | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | 登録日 | 2001-04-12 | | 公開日 | 2001-05-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LU0
 
 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | 登録日 | 2002-05-21 | | 公開日 | 2002-08-28 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | 分子名称: | NEURAL CELL ADHESION MOLECULE | | 著者 | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | 登録日 | 2001-04-06 | | 公開日 | 2001-08-08 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1J4W
 
 | |
1LZ8
 
 | | LYSOZYME PHASED ON ANOMALOUS SIGNAL OF SULFURS AND CHLORINES | | 分子名称: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Dauter, Z, Dauter, M, De La Fortelle, E, Bricogne, G, Sheldrick, G.M. | | 登録日 | 1999-03-14 | | 公開日 | 1999-05-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Can anomalous signal of sulfur become a tool for solving protein crystal structures?
J.Mol.Biol., 289, 1999
|
|
1MIE
 
 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | 分子名称: | IMMUNOGLOBULIN MS5-393 | | 著者 | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | 登録日 | 2002-08-23 | | 公開日 | 2003-09-23 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJJ
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | 分子名称: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | 著者 | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | 登録日 | 2002-08-28 | | 公開日 | 2003-09-23 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1M32
 
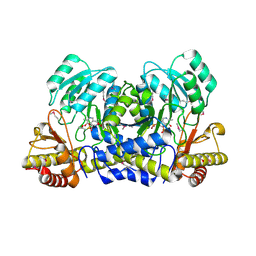 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | 分子名称: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | 著者 | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | 登録日 | 2002-06-26 | | 公開日 | 2002-11-20 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
7DJJ
 
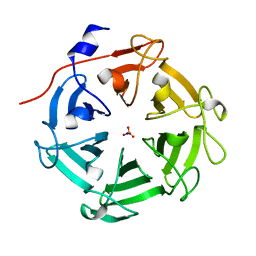 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | 分子名称: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.69806433 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
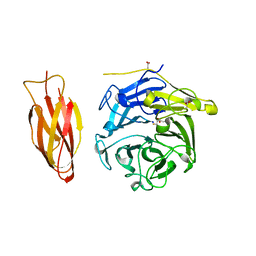 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.70000112 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
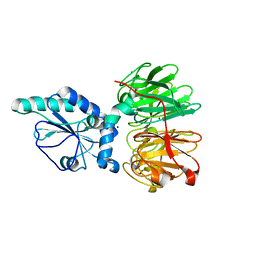 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.80145121 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.96077824 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7I1B
 
 | |
2HEQ
 
 | | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399. | | 分子名称: | YorP protein | | 著者 | Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.M, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-06-21 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399.
To be Published
|
|
1EMV
 
 | | CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS) | | 分子名称: | COLICIN E9, IMMUNITY PROTEIN IM9, PHOSPHATE ION | | 著者 | Kuhlmann, U.C, Pommer, A.J, Moore, G.M, James, R, Kleanthous, C. | | 登録日 | 2000-03-20 | | 公開日 | 2000-09-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes.
J.Mol.Biol., 301, 2000
|
|
1FEU
 
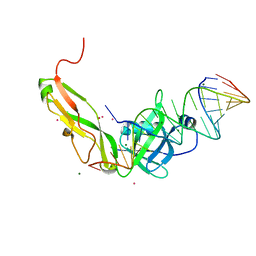 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | 分子名称: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | 著者 | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | 登録日 | 2000-07-23 | | 公開日 | 2001-06-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GIC
 
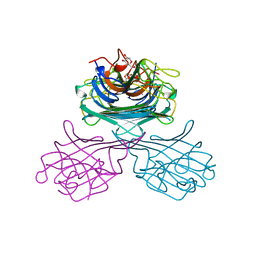 | | CONCANAVALIN A COMPLEXED WITH METHYL ALPHA-D-GLUCOPYRANOSIDE | | 分子名称: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | 著者 | Bradbrook, G.M, Gleichmann, T, Harrop, S.J, Helliwell, J.R, Habash, J, Kalb(Gilboa), A.J, Tong, L, Wan, T.C.M, Yariv, J. | | 登録日 | 1996-08-08 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure solution of a cubic crystal of concanavalin A complexed with methyl alpha-D-glucopyranoside.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7JGC
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase FO region | | 分子名称: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG9
 
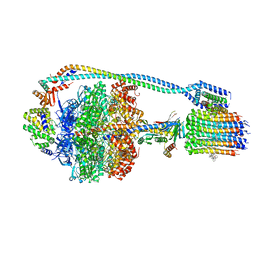 | | Cryo-EM structure of bedaquiline-saturated mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
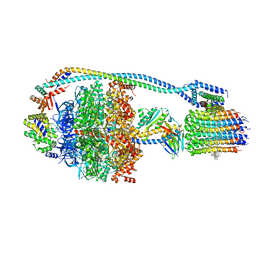 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGB
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase FO region | | 分子名称: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG6
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
