2CDP
 
 | | Structure of a CBM6 in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2C8N
 
 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,3-linked arabinoside of xylobiose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-3)-alpha-D-xylopyranose | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
6GYI
 
 | |
2C01
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2BO4
 
 | | Dissection of mannosylglycerate synthase: an archetypal mannosyltransferase | | Descriptor: | CITRATE ANION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
6FHW
 
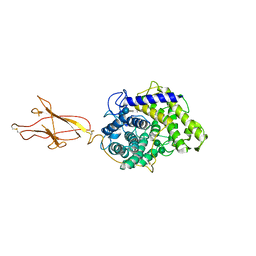 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2BLN
 
 | | N-terminal formyltransferase domain of ArnA in complex with N-5- formyltetrahydrofolate and UMP | | Descriptor: | ACETATE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PROTEIN YFBG, ... | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L-Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
6FHJ
 
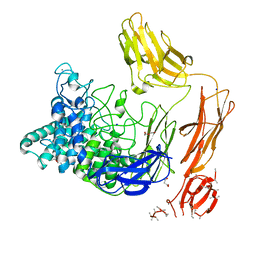 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
2CIT
 
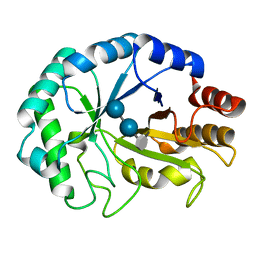 | | Structure of the covalent intermediate of a family 26 lichenase | | Descriptor: | ENDOGLUCANASE H, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Money, V.A, Smith, N.L, Scaffidi, A, Stick, R.V, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate Distortion by a Lichenase Highlights the Different Conformational Itineraries Harnessed by Related Glycoside Hydrolases.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2C7F
 
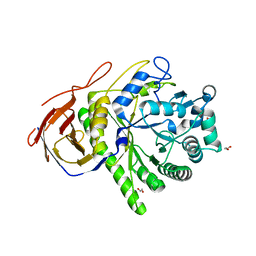 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,5-alpha-L-Arabinotriose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
6G3E
 
 | | Crystal structure of EDDS lyase in complex with formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
2CDS
 
 | | LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME (E.C.3.2.1.17)) | | Authors: | Kleywegt, G.J, Divne, C. | | Deposit date: | 1999-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Lysozyme
To be Published
|
|
2CIP
 
 | | Structure of the Michaelis complex of a family 26 lichenase | | Descriptor: | 4-METHYL-2H-CHROMEN-2-ONE, ENDOGLUCANASE H, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Money, V.A, Smith, N.L, Scaffidi, A, Stick, R.V, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate Distortion by a Lichenase Highlights the Different Conformational Itineraries Harnessed by Related Glycoside Hydrolases.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
6G0B
 
 | |
2CHO
 
 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
6G09
 
 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G1I
 
 | | GH124 cellulase from Ruminiclostridium thermocellum in complex with Mn and fructosylated cellopentaose | | Descriptor: | Glycosyl Hydrolase, MALONIC ACID, MANGANESE (II) ION, ... | | Authors: | Urresti, S, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural studies of the unusual metal-ion site of the GH124 endoglucanase from Ruminiclostridium thermocellum.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6GXV
 
 | | Amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, A-amylase, CALCIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GYA
 
 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2C3Q
 
 | | Human glutathione-S-transferase T1-1 W234R mutant, complex with S- hexylglutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2BVD
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
6G96
 
 | | Crystal structure of TacT3 (tRNA acetylating toxin) from Salmonella | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, BICINE, ... | | Authors: | Grabe, G.J, Rycroft, J.A, Gollan, B, Hall, A, Cheverton, A.M, Larrouy-Maumus, G, Hare, S.A, Helaine, S. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4766078 Å) | | Cite: | Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection.
Nat Commun, 9, 2018
|
|
2C11
 
 | | Crystal structure of the 2-hydrazinopyridine of semicarbazide- sensitive amine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jakobsson, E, Kleywegt, G.J. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human semicarbazide-sensitive amine oxidase/vascular adhesion protein-1.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
6G21
 
 | | Crystal structure of an esterase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an esterase from Aspergillus oryzae
To Be Published
|
|
6G1G
 
 | |
