5N7B
 
 | | Understanding the singular conformational landscape of the Tn antigens: Sulfur-for- oxygen substitution in the glycosidic linkage provides new insights into molecular recognition by an antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, APD(CG6)RP(NH2) peptide, ... | | Authors: | Companon, I, Martinez-Saez, N, Castro-Lopez, J, Jimenez-Barbero, J, Bernardes, G.J.L, Busto, J.H, Avenoza, A, Jimenez-Oses, G, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2017-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of Potent Tumor-Associated Antigens: Modulation of Peptide Presentation by Single-Atom O/S or O/Se Substitutions at the Glycosidic Linkage.
J.Am.Chem.Soc., 141, 2019
|
|
5O1D
 
 | | p53 cancer mutant Y220C in complex with compound MB481 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-propoxy-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1C
 
 | | p53 cancer mutant Y220C in complex with compound MB184 | | Descriptor: | 5-(4-fluorophenyl)-3-iodanyl-2-oxidanyl-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1I
 
 | | p53 cancer mutant Y220C in complex with compound MB710 | | Descriptor: | 1,2-ETHANEDIOL, 2-(diethylamino)-6-iodanyl-5-oxidanyl-7-pyrrol-1-yl-1,3-benzothiazole-4-carboxylic acid, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1A
 
 | | p53 cancer mutant Y220C in complex with compound MB240 | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1H
 
 | | p53 cancer mutant Y220C in complex with compound MB539 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-propylsulfanyl-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1F
 
 | | p53 cancer mutant Y220C in complex with compound MB582 | | Descriptor: | 5-butoxy-3-iodanyl-2-oxidanyl-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5N7V
 
 | | TTK kinase domain in complex with MPI-0479605 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, ~{N}6-cyclohexyl-~{N}2-(2-methyl-4-morpholin-4-yl-phenyl)-7~{H}-purine-2,6-diamine | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, Zaman, G.J.R. | | Deposit date: | 2017-02-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5O1G
 
 | | p53 cancer mutant Y220C in complex with compound MB487 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-(2-phenylethoxy)-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1B
 
 | | p53 cancer mutant Y220C in complex with compound MB84 | | Descriptor: | 6-(hydroxymethyl)-2,4-bis(iodanyl)-3-pyrrol-1-yl-phenol, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
2QV8
 
 | |
2I4S
 
 | | PDZ domain of EpsC from Vibrio cholerae, residues 204-305 | | Descriptor: | General secretion pathway protein C | | Authors: | Korotkov, K.V, Krumm, B, Bagdasarian, M, Hol, W.G.J. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
J.Mol.Biol., 363, 2006
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2RET
 
 | | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the Type 2 Secretion System of Vibrio vulnificus | | Descriptor: | CHLORIDE ION, EpsJ, Pseudopilin EpsI, ... | | Authors: | Yanez, M.E, Korotkov, K.V, Abendroth, J, Hol, W.G.J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
J.Mol.Biol., 375, 2008
|
|
2TEC
 
 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2SBT
 
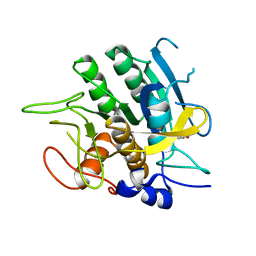 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
2VJO
 
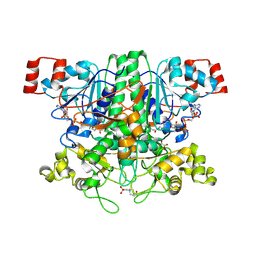 | | Formyl-CoA transferase mutant variant Q17A with aspartyl-CoA thioester intermediates and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, COENZYME A, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VJL
 
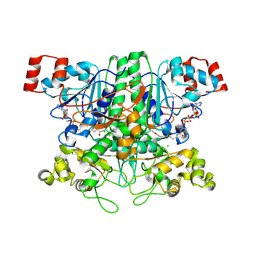 | | Formyl-CoA transferase with aspartyl-CoA thioester intermediate derived from formyl-CoA | | Descriptor: | CHLORIDE ION, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VMB
 
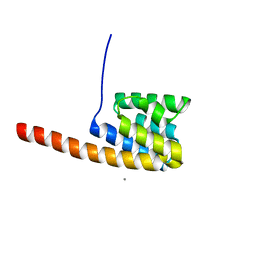 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2IV9
 
 | |
2IV8
 
 | |
2VMA
 
 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F, IODIDE ION | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2VJN
 
 | | Formyl-CoA transferase mutant variant G260A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMYL-COENZYME A TRANSFERASE | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2JI8
 
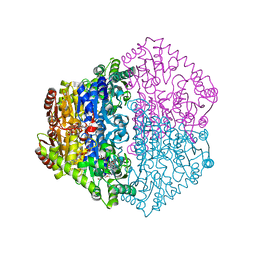 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with Formyl- CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OXALYL-COA DECARBOXYLASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2W7V
 
 | | periplasmic domain of EpsL from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, GENERAL SECRETION PATHWAY PROTEIN L, PHOSPHATE ION | | Authors: | Abendroth, J, Kreger, A.C, Abendroth, H, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2009-01-06 | | Release date: | 2010-03-31 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Dimer Formed by the Periplasmic Domain of Epsl from the Type 2 Secretion System of Vibrio Parahaemolyticus.
J.Struct.Biol., 168, 2009
|
|
