3BUE
 
 | | Crystal structure of the C-terminal domain hexamer of ArgR from Mycobacterium tuberculosis | | Descriptor: | Arginine repressor ArgR | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HTI
 
 | |
1HI2
 
 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
6DI7
 
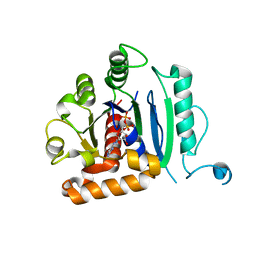 | | Vps1 GTPase-BSE fusion complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative sorting protein | | Authors: | Varlakhanova, N.V, Brady, T.M, Ford, M.G.J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
1I5J
 
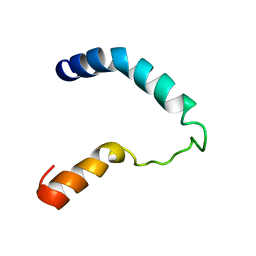 | |
1IIG
 
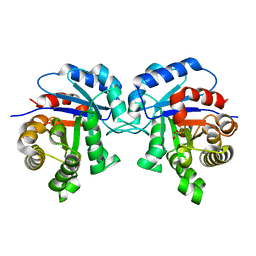 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
1INQ
 
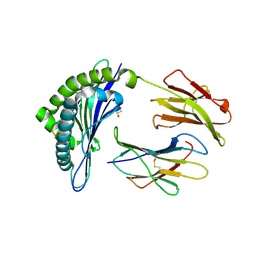 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | Descriptor: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
1IIH
 
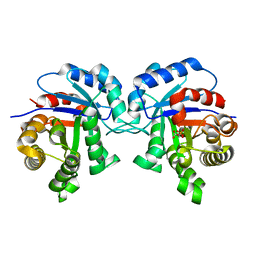 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
5AQE
 
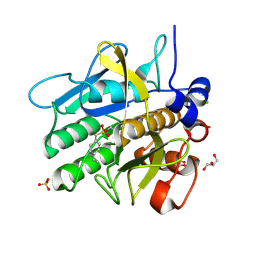 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | (4-VINYLPHENYL)METHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bisubstrate Localisation by a Tailored Serine Protease Allows Creation of a Heck-Ase
To be Published
|
|
3VPS
 
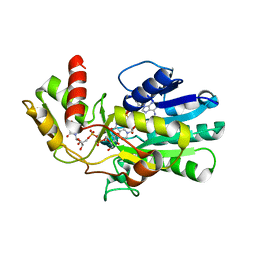 | | Structure of a novel NAD dependent-NDP-hexosamine 5,6-dehydratase, TunA, involved in tunicamycin biosynthesis | | Descriptor: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Wyszynski, F.J, Lee, S.S, Yabe, T, Wang, H, Gomez-Escribano, J.P, Bibb, M.J, Lee, S.J, Davies, G.J, Davis, B.G. | | Deposit date: | 2012-03-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biosynthesis of nucleoside antibiotic tunicamycin proceeds via unique exo-glycal intermediates
To be published
|
|
1JW0
 
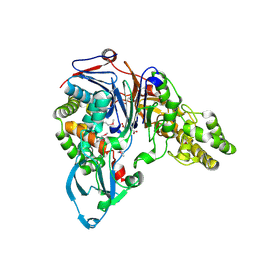 | | Structure of cephalosporin acylase in complex with glutarate | | Descriptor: | GLUTARIC ACID, cephalosporin acylase alpha chain, cephalosporin acylase beta chain | | Authors: | Kim, Y, Hol, W.G.J. | | Deposit date: | 2001-09-01 | | Release date: | 2002-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid and glutarate: insight into the basis of its substrate specificity
CHEM.BIOL., 8, 2001
|
|
5ARB
 
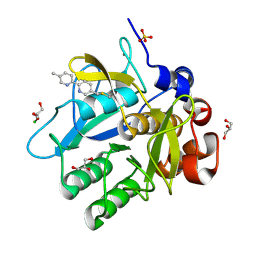 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5CAX
 
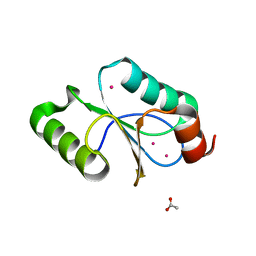 | | CRYSTAL STRUCTURE OF METHANOSARCINA ACETIVORANS METHANOREDOXIN | | Descriptor: | ACETIC ACID, CADMIUM ION, Glutaredoxin, ... | | Authors: | Yennawar, N.H, Yennawar, H.P, Ferry, G.J. | | Deposit date: | 2015-06-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural and Biochemical Characterizations of Methanoredoxin from Methanosarcina acetivorans, a Glutaredoxin-Like Enzyme with Coenzyme M-Dependent Protein Disulfide Reductase Activity.
Biochemistry, 55, 2016
|
|
1H6X
 
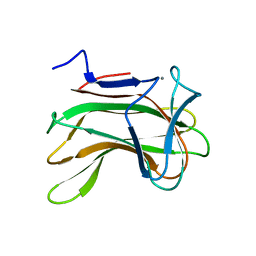 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1HFA
 
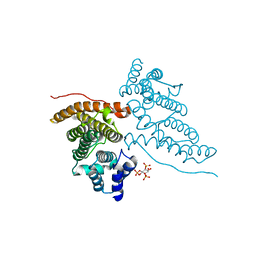 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, PI(4,5)P2 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-11-30 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
1HFK
 
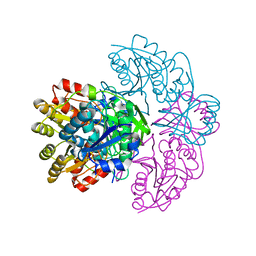 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with weak sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Lubkowski, J, Palm, G.J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H7L
 
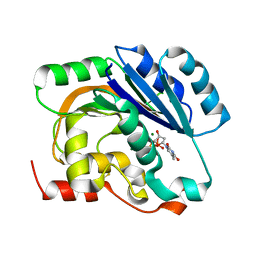 | | dTDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
5C8V
 
 | |
1HB8
 
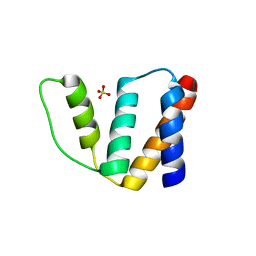 | | Structure of bovine Acyl-CoA binding protein in tetragonal crystal form | | Descriptor: | ACYL-COA BINDING PROTEIN, SULFATE ION | | Authors: | Zou, J.Y, Kleywegt, G.J, Bergfors, T, Knudsen, J, Jones, T.A. | | Deposit date: | 2001-04-12 | | Release date: | 2002-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Site Differences Revealed by Crystal Structures of Plasmodium Falciparum and Bovine Acyl-Coa Binding Protein
J.Mol.Biol., 309, 2001
|
|
5CA5
 
 | | Structure of the C. elegans NONO-1 homodimer | | Descriptor: | 1,2-ETHANEDIOL, NONO-1 | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2015-06-29 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caenorhabditis elegans NONO-1: Insights into DBHS protein structure, architecture, and function.
Protein Sci., 24, 2015
|
|
1H0A
 
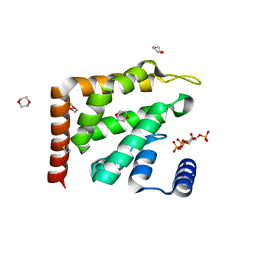 | | Epsin ENTH bound to Ins(1,4,5)P3 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, EPSIN | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2002-06-12 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Curvature of Clathrin-Coated Pits Driven by Epsin
Nature, 419, 2002
|
|
1H7Q
 
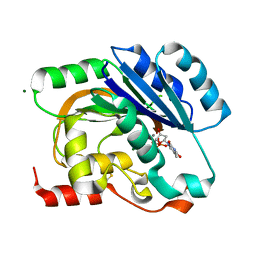 | | dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, ... | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
