6KXD
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6LCE
 
 | |
5YHS
 
 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, apo form | | Descriptor: | Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YB7
 
 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-ornithine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase, L-ornithine | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YIF
 
 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YB6
 
 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-lysine complex | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase, ... | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YSD
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YB8
 
 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-arginine complex | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
6A3F
 
 | | Levoglucosan dehydrogenase, apo form | | Descriptor: | Putative dehydrogenase, SULFATE ION | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A3I
 
 | | Levoglucosan dehydrogenase, complex with NADH and levoglucosan | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Levoglucosan, Putative dehydrogenase | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A3G
 
 | | Levoglucosan dehydrogenase, complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative dehydrogenase | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5XB7
 
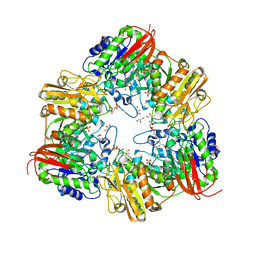 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
3UG4
 
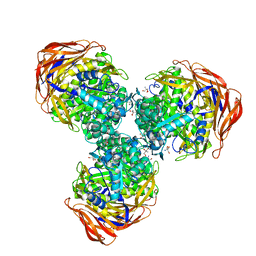 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
6A3J
 
 | | Levoglucosan dehydrogenase, complex with NADH and L-sorbose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative dehydrogenase, ... | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
3UG5
 
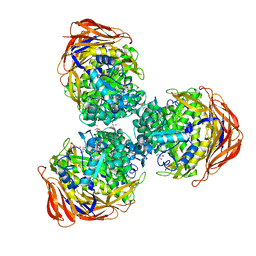 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UG3
 
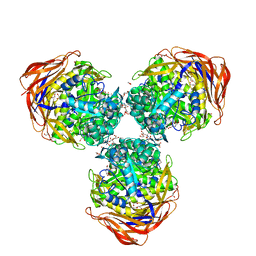 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3AO9
 
 | | Crystal structure of the C-terminal domain of sequence-specific ribonuclease | | Descriptor: | CADMIUM ION, Colicin-E5 | | Authors: | Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H, Yajima, S. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
3X2N
 
 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2Z8F
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-tetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2Z8D
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2DTJ
 
 | | Crystal structure of regulatory subunit of aspartate kinase from Corynebacterium glutamicum | | Descriptor: | Aspartokinase, CITRIC ACID, THREONINE | | Authors: | Yoshida, A, Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2006-07-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Insight into Concerted Inhibition of alpha(2)beta(2)-Type Aspartate Kinase from Corynebacterium glutamicum
J.Mol.Biol., 368, 2007
|
|
3AC0
 
 | | Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose | | Descriptor: | Beta-glucosidase I, beta-D-glucopyranose | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
