1BK1
 
 | | ENDO-1,4-BETA-XYLANASE C | | 分子名称: | ENDO-1,4-B-XYLANASE C | | 著者 | Fushinobu, S, Ito, K, Konno, M, Wakagi, T, Matsuzawa, H. | | 登録日 | 1998-07-14 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH.
Protein Eng., 11, 1998
|
|
3R1M
 
 | | Structure of bifunctional fructose 1,6-bisphosphate aldolase/phosphatase (aldolase form) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Fushinobu, S, Nishimasu, H, Hattori, D, Song, H.-J, Wakagi, T. | | 登録日 | 2011-03-10 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
8HVB
 
 | |
8HVC
 
 | |
8HVD
 
 | |
3QG0
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin | | 分子名称: | 1-DEOXYNOJIRIMYCIN, Cellobiose Phosphorylase, PHOSPHATE ION, ... | | 著者 | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2011-01-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3RI9
 
 | | Xylanase C from Aspergillus kawachii F131W mutant | | 分子名称: | Endo-1,4-beta-xylanase 3 | | 著者 | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | 登録日 | 2011-04-13 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3RI8
 
 | | Xylanase C from Aspergillus kawachii D37N mutant | | 分子名称: | Endo-1,4-beta-xylanase 3 | | 著者 | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | 登録日 | 2011-04-13 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
1WU5
 
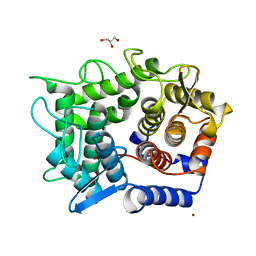 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase complexed with xylose | | 分子名称: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose, ... | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU4
 
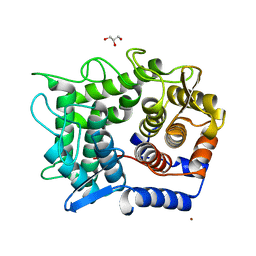 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | 分子名称: | GLYCEROL, NICKEL (II) ION, xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU6
 
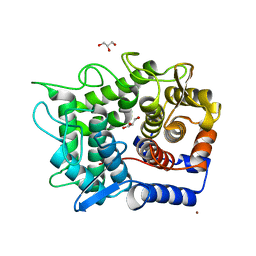 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase E70A mutant complexed with xylobiose | | 分子名称: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
3QFZ
 
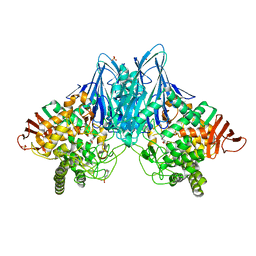 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin | | 分子名称: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiose Phosphorylase, ... | | 著者 | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2011-01-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QFY
 
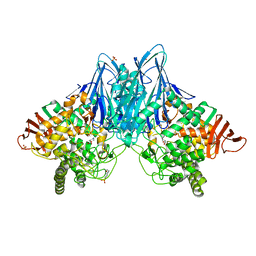 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Cellobiose Phosphorylase, ... | | 著者 | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2011-01-24 | | 公開日 | 2011-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
2DRR
 
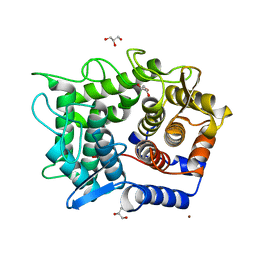 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRS
 
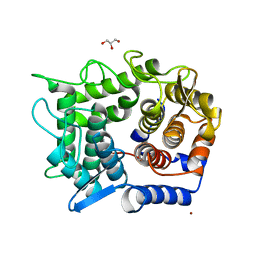 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263S mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRO
 
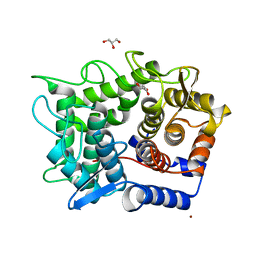 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263C mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRQ
 
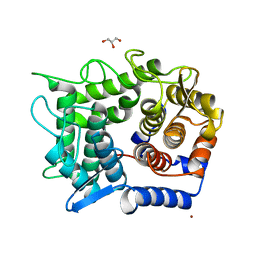 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263G mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DEP
 
 | |
1J30
 
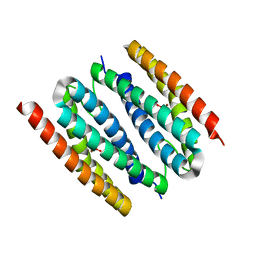 | | The crystal structure of sulerythrin, a rubrerythrin-like protein from a strictly aerobic and thermoacidiphilic archaeon | | 分子名称: | 144aa long hypothetical rubrerythrin, FE (III) ION, OXYGEN MOLECULE, ... | | 著者 | Fushinobu, S, Shoun, H, Wakagi, T. | | 登録日 | 2003-01-16 | | 公開日 | 2003-10-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of Sulerythrin, A Rubrerythrin-like Protein from A Strictly Aerobic Archaeon, Sulfolobus tokodaii strain 7, shows unexpected domain swapping
Biochemistry, 42, 2003
|
|
1IUP
 
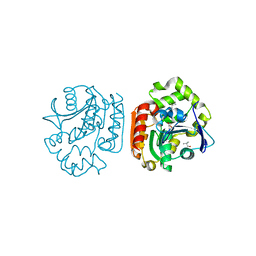 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with isobutyrates | | 分子名称: | 2-METHYL-PROPIONIC ACID, meta-Cleavage product hydrolase | | 著者 | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2002-03-06 | | 公開日 | 2002-09-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1IUN
 
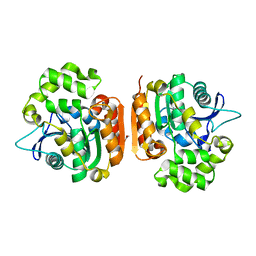 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant hexagonal | | 分子名称: | ACETATE ION, meta-Cleavage product hydrolase | | 著者 | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2002-03-06 | | 公開日 | 2002-09-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1IUO
 
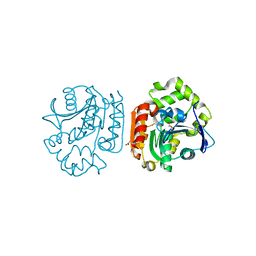 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with acetates | | 分子名称: | ACETATE ION, meta-Cleavage product hydrolase | | 著者 | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2002-03-06 | | 公開日 | 2002-09-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1UK6
 
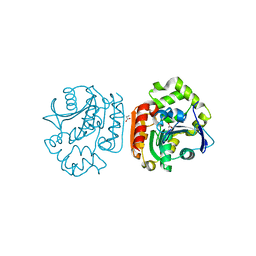 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with propionate | | 分子名称: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PROPANOIC ACID | | 著者 | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2003-08-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK8
 
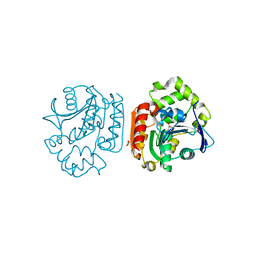 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-valerate | | 分子名称: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PENTANOIC ACID | | 著者 | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2003-08-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK7
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate | | 分子名称: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, butanoic acid | | 著者 | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | 登録日 | 2003-08-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
