5H42
 
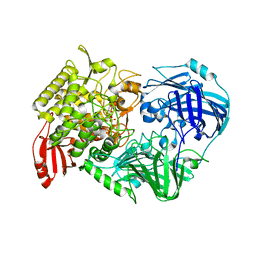 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
4KTP
 
 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with glucose | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 65 central catalytic, PENTAETHYLENE GLYCOL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
5H41
 
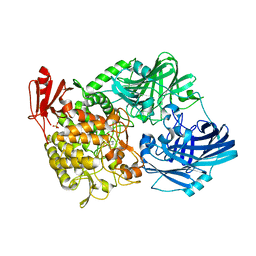 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
6KQS
 
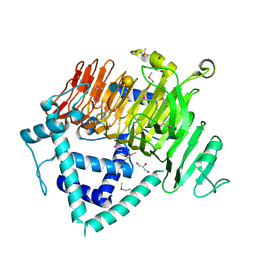 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6LCE
 
 | |
1X0L
 
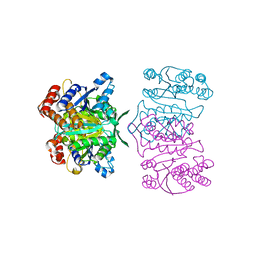 | | Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus | | Descriptor: | Homoisocitrate dehydrogenase | | Authors: | Miyazaki, J, Asada, K, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-03-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetrameric Homoisocitrate Dehydrogenase from an Extreme Thermophile, Thermus thermophilus: Involvement of Hydrophobic Dimer-Dimer Interaction in Extremely High Thermotolerance
J.Bacteriol., 187, 2005
|
|
1WD3
 
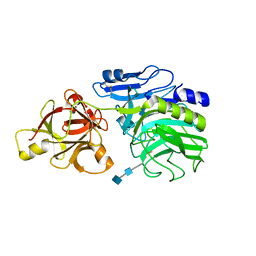 | | Crystal structure of arabinofuranosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1WD4
 
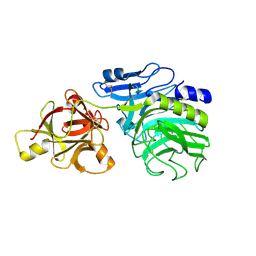 | | Crystal structure of arabinofuranosidase complexed with arabinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1XQD
 
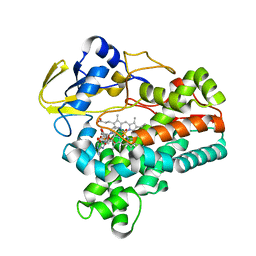 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
4KTR
 
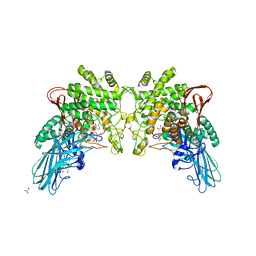 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with isofagomine and glycerol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
7CL9
 
 | |
7CL7
 
 | |
5H3Z
 
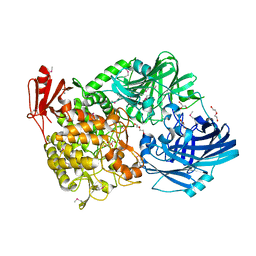 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
1VCK
 
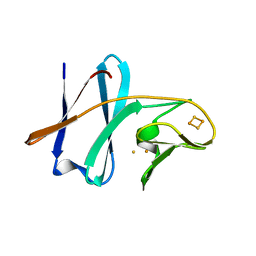 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
4JAW
 
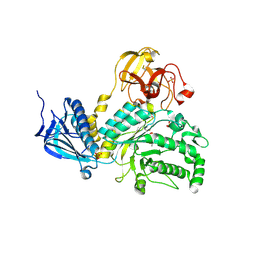 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
6KQT
 
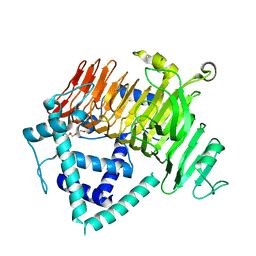 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
1J2A
 
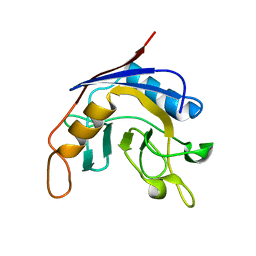 | | Structure of E. coli cyclophilin B K163T mutant | | Descriptor: | cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2002-12-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
5GZK
 
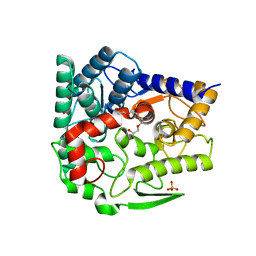 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
4ZLG
 
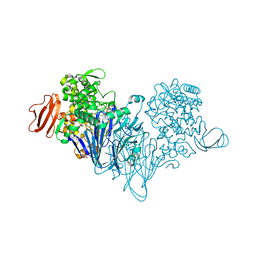 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLF
 
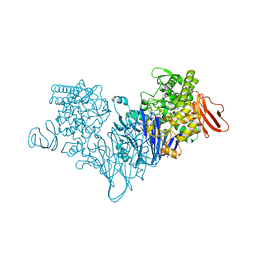 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
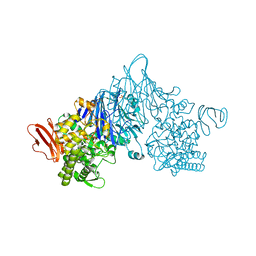 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | Descriptor: | (SIN)APA(NIT), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
