2Z8K
 
 | |
2DBU
 
 | |
1I7H
 
 | | CRYSTAL STURCUTURE OF FDX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Kakuta, Y, Horio, T, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2001-03-09 | | Release date: | 2002-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli Fdx, an adrenodoxin-type ferredoxin involved in the assembly of iron-sulfur clusters.
Biochemistry, 40, 2001
|
|
3A75
 
 | |
3AJH
 
 | | Crystal structure of PcyA V225D-biliverdin XIII alpha complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Wada, K, Hagiwara, Y, Fukuyama, K. | | Deposit date: | 2010-06-05 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | One residue substitution in PcyA leads to unexpected changes in tetrapyrrole substrate binding.
Biochem.Biophys.Res.Commun., 402, 2010
|
|
3AJG
 
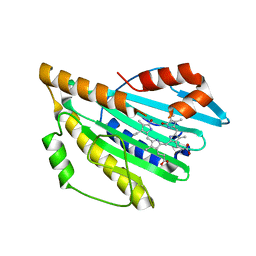 | |
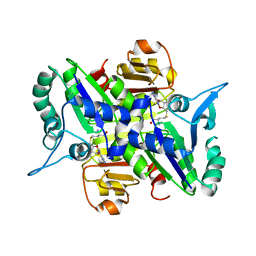
2E0N
 
 | |
2E0K
 
 | |
2D2A
 
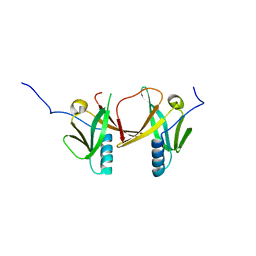 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
2D3W
 
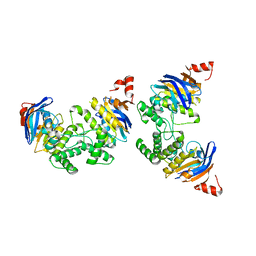 | | Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery | | Descriptor: | Probable ATP-dependent transporter sufC | | Authors: | Kitaoka, S, Wada, K, Hasegawa, Y, Minami, Y, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery
Febs Lett., 580, 2006
|
|
2E7E
 
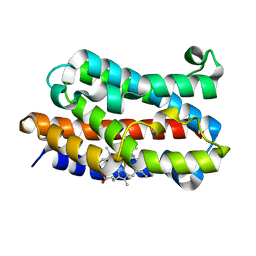 | |
2ZVU
 
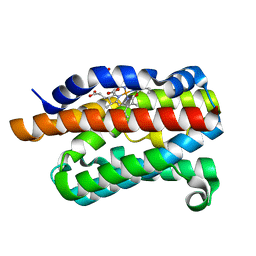 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
2ZXL
 
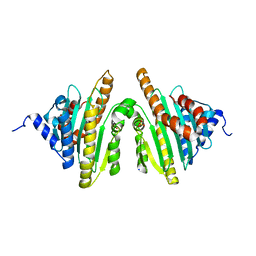 | | Crystal structure of red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | Red chlorophyll catabolite reductase, chloroplastic, SODIUM ION, ... | | Authors: | Sugishima, M, Kitamori, Y, Fukuyama, K. | | Deposit date: | 2008-12-29 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of red chlorophyll catabolite reductase: enlargement of the ferredoxin-dependent bilin reductase family
J.Mol.Biol., 389, 2009
|
|
3AGC
 
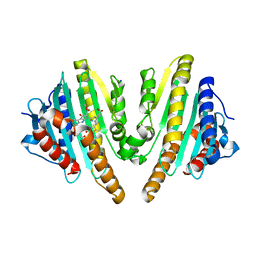 | | F218V mutant of the substrate-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGA
 
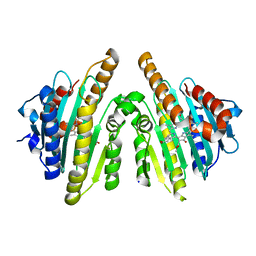 | | Crystal structure of RCC-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGB
 
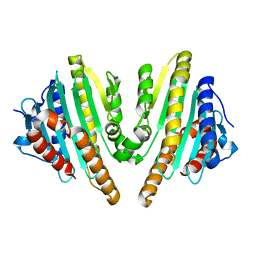 | |
2ZXK
 
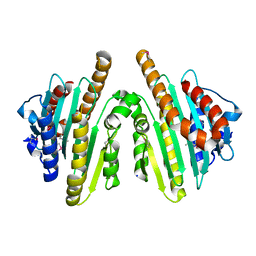 | |
2D1E
 
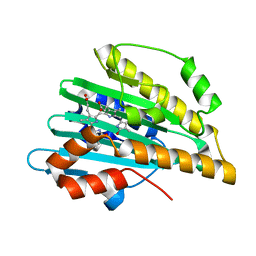 | | Crystal structure of PcyA-biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DKE
 
 | | Crystal structure of substrate-free form of PcyA | | Descriptor: | CHLORIDE ION, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Induced-fitting and electrostatic potential change of PcyA upon substrate binding demonstrated by the crystal structure of the substrate-free form
Febs Lett., 580, 2006
|
|
2DY5
 
 | | Crystal structure of rat heme oxygenase-1 in complex with heme and 2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-1,3-dioxolane | | Descriptor: | 1-({2-[2-(4-CHLOROPHENYL)ETHYL]-1,3-DIOXOLAN-2-YL}METHYL)-1H-IMIDAZOLE, CHLORIDE ION, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Takahashi, H, Fukuyama, K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic and biochemical characterization of the inhibitory action of an imidazole-dioxolane compound on heme oxygenase
Biochemistry, 46, 2007
|
|
