3I95
 
 | |
1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
3WHQ
 
 | |
3WHS
 
 | | Crystal structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase large chain, Gamma-glutamyltranspeptidase small chain | | Authors: | Wada, K, Fukuyama, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WHR
 
 | | Crystal structure of gamma-glutamyltranspeptidase from Bacillus subtilis (crystal soaked for 3min. in acivicin soln. ) | | Descriptor: | Gamma-glutamyltranspeptidase large chain, Gamma-glutamyltranspeptidase small chain | | Authors: | Ida, T, Suzuki, H, Fukuyama, K, Hiratake, J, Wada, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKT
 
 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1D2M
 
 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
1DVE
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
5B4I
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 2) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5B4H
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 1) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5B4J
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 3) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
1IRM
 
 | | Crystal structure of apo heme oxygenase-1 | | Descriptor: | apo heme oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Kakuta, Y, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2001-10-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of rat apo-heme oxygenase-1 (HO-1): mechanism of heme binding in HO-1 inferred from structural comparison of the apo and heme complex forms
Biochemistry, 41, 2002
|
|
5B5T
 
 | | Crystal Structure of Escherichia coli Gamma-Glutamyltranspeptidase in Complex with peptidyl phosphonate inhibitor 1b | | Descriptor: | (2~{S})-2-azanyl-4-[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-butan-2-yl]oxyphosphonoyl-butanoic acid, CALCIUM ION, Gamma-glutamyltranspeptidase large chain, ... | | Authors: | Wada, K, Fukuyama, K. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Phosphonate-based irreversible inhibitors of human gamma-glutamyl transpeptidase (GGT). GGsTop is a non-toxic and highly selective inhibitor with critical electrostatic interaction with an active-site residue Lys562 for enhanced inhibitory activity
Bioorg.Med.Chem., 24, 2016
|
|
1UGU
 
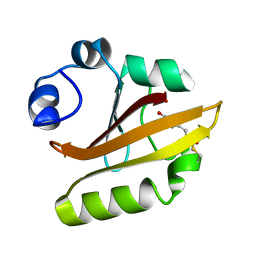 | | Crystal structure of PYP E46Q mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Sugishima, M, Tanimoto, Y, Hamada, N, Tokunaga, F, Fukuyama, K. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ARP
 
 | |
1UBB
 
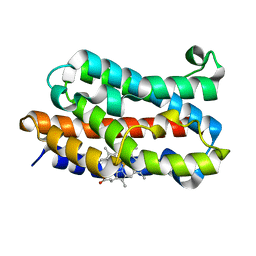 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1ULX
 
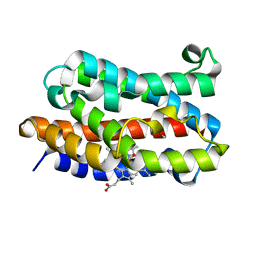 | | Partially photolyzed structure of CO-bound heme-heme oxygenase complex | | Descriptor: | CARBON MONOXIDE, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-09-16 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
1IZ0
 
 | |
1IYZ
 
 | |
1UJ8
 
 | |
1IX3
 
 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Cyanide | | Descriptor: | CYANIDE ION, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1IX4
 
 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Carbon Monoxide | | Descriptor: | CARBON MONOXIDE, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-06-10 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
1X1D
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn-bacteriopheophorbide d | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-04 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1B
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
