1HVX
 
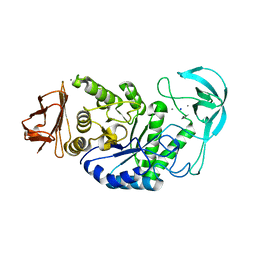 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
7YL5
 
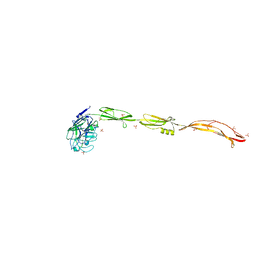 | | Cell surface protein YwfG protein complexed with mannose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
1UA7
 
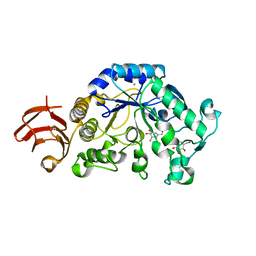 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
7YL6
 
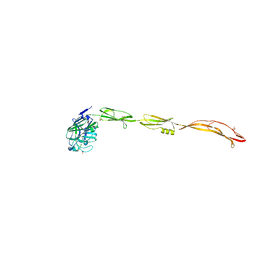 | | Cell surface protein YwfG protein complexed with alpha-1,2-mannobiose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL4
 
 | | Cell surface protein YwfG protein (apo form) | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
2G3I
 
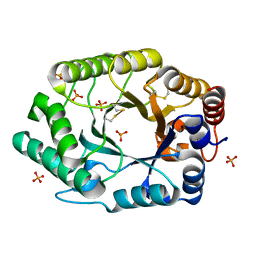 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
1J35
 
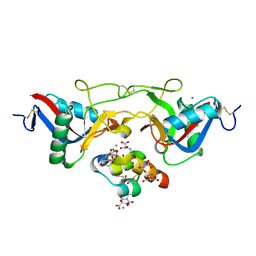 | | Crystal Structure of Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, Coagulation factor IX-binding protein B chain, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
1J34
 
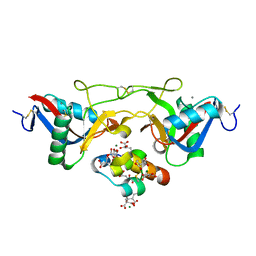 | | Crystal Structure of Mg(II)-and Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, MAGNESIUM ION, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
5GQE
 
 | | Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
5GQD
 
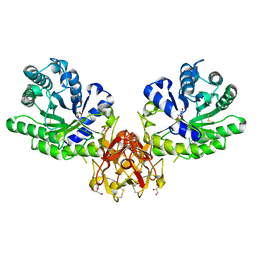 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
1J2L
 
 | | Crystal structure of the disintegrin, trimestatin | | Descriptor: | Disintegrin triflavin, SULFATE ION | | Authors: | Fujii, Y, Okuda, D, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-10-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Trimestatin, a Disintegrin Containing a Cell Adhesion Recognition Motif RGD
J.Mol.Biol., 332, 2003
|
|
5AXH
 
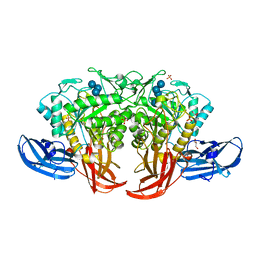 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
5AXG
 
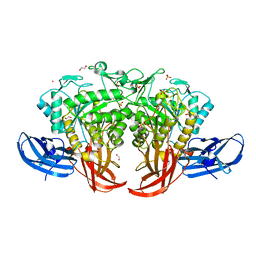 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Dextranase, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
2D24
 
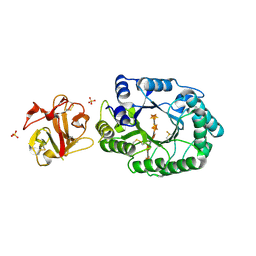 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
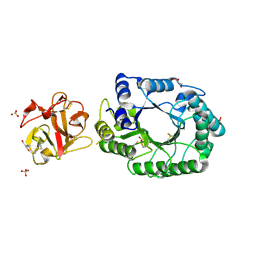 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D1Z
 
 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
1IOD
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE COAGULATION FACTOR X BINDING PROTEIN FROM SNAKE VENOM AND THE GLA DOMAIN OF FACTOR X | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X BINDING PROTEIN, COAGULATION FACTOR X GLA DOMAIN | | Authors: | Mizuno, H, Fujimoto, Z, Atoda, H, Morita, T. | | Deposit date: | 2001-02-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an anticoagulant protein in complex with the Gla domain of factor X.
Proc.Natl.Acad.Sci.Usa, 98, 2001
|
|
2DS0
 
 | |
2DRY
 
 | |
2DRZ
 
 | |
5GLK
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome, calcium-free form. | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLO
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose, calcium-free form | | Descriptor: | ACETATE ION, Glycoside hydrolase family 43, SODIUM ION, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLQ
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose and xylotriose, calcium-free form | | Descriptor: | Glycoside hydrolase family 43, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
