3DTD
 
 | | Crystal structure of invasion associated protein b from bartonella henselae | | Descriptor: | GLYCEROL, Invasion-associated protein B | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Slocombe, A, Groshong, C, Koss, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Invasion Associated Protein B from Bartonella Henselae.
To be Published
|
|
3ESM
 
 | | Crystal structure of an uncharacterized protein from Nocardia farcinica reveals an immunoglobulin-like fold | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, uncharacterized protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an uncharacterized protein from Nocardia farcinica reveals an immunoglobulin-like fold
To be Published
|
|
3DBI
 
 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3DBY
 
 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
3FI9
 
 | | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis | | Descriptor: | Malate dehydrogenase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis
To be Published
|
|
3C8C
 
 | | Crystal structure of Mcp_N and cache domains of methyl-accepting chemotaxis protein from Vibrio cholerae | | Descriptor: | ALANINE, MAGNESIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Hu, S, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-19 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae.
To be Published
|
|
3CRN
 
 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, Response regulator receiver domain protein, CheY-like, ... | | Authors: | Hiner, R.L, Toro, R, Patskovsky, Y, Freeman, J, Chang, S, Smith, D, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1.
To be Published
|
|
3DO9
 
 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
3CYM
 
 | | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein BAD_0989 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Chang, S, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis.
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3CG4
 
 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Response regulator receiver domain protein (CheY-like) | | Authors: | Patskovsky, Y, Freeman, J, Hu, S, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Response Regulator Receiver Domain Protein (Chey-Like) from Methanospirillum hungatei JF-1.
To be Published
|
|
3CZB
 
 | | Crystal structure of putative transglycosylase from Caulobacter crescentus | | Descriptor: | Putative transglycosylase, SULFATE ION | | Authors: | Ramagopal, U.A, Chattopadhyay, K, Toro, R, Wasserman, S, Freeman, J, Logan, C, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative transglycosylase from Caulobacter crescentus.
To be Published
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | Descriptor: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
3GHY
 
 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3BE7
 
 | | Crystal structure of Zn-dependent arginine carboxypeptidase | | Descriptor: | ARGININE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IQ1
 
 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2HY3
 
 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3PVE
 
 | | Crystal structure of the G2 domain of Agrin from Mus Musculus | | Descriptor: | Agrin, Agrin protein | | Authors: | Sampathkumar, P, Do, J, Bain, K, Freeman, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the G2 domain of Agrin from Mus Musculus
To be Published
|
|
2FH7
 
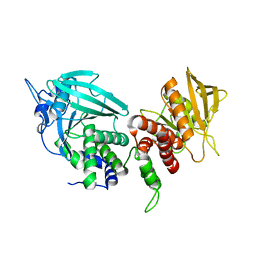 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3MN1
 
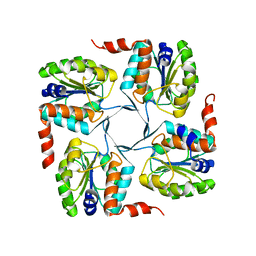 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
