7NL3
 
 | |
7NDE
 
 | | Trichoderma parareesei PL7A beta-glucuronan lyase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucuronan lyase | | Authors: | Fredslund, F, Welner, D.H, Wilkens, C. | | Deposit date: | 2021-02-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glucuronan lyases from family PL7 use a Tyr/Tyr syn beta-elimination catalytic mechanism for glucuronan breakdown.
Chem.Commun.(Camb.), 60, 2024
|
|
7NCZ
 
 | |
7NPP
 
 | |
7NY3
 
 | |
7O6H
 
 | |
6RZN
 
 | |
7OSK
 
 | | Ignisphaera aggregans GH53 catalytic domain | | Descriptor: | Arabinogalactan endo-1,4-beta-galactosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fredslund, F, Lo Leggio, L, Poulsen, J.C, Rasmussen, K.K, Muderspach, S, Krogh, K.B.R.M, Jensen, K. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering the substrate binding site of the hyperthermostable archaeal endo-beta-1,4-galactanase from Ignisphaera aggregans.
Biotechnol Biofuels, 14, 2021
|
|
7P90
 
 | |
6RZO
 
 | |
7PBF
 
 | |
7OOF
 
 | |
7ORY
 
 | |
7P25
 
 | |
6SU7
 
 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline | | Descriptor: | 3,4-Dichloroaniline, Glycosyltransferase | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
6SU6
 
 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and UDP-glucose | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
3KLS
 
 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KM9
 
 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7ZF0
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZER
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
6YJI
 
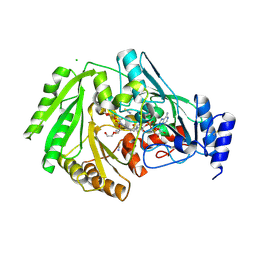 | | Structure of FgCelDH7C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Haddad Momeni, M, Fredslund, F, Berrin, J.G, Abou Hachem, M, Welner, D.H. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs.
Nat Commun, 12, 2021
|
|
6YJO
 
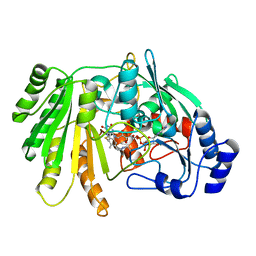 | | Structure of FgChi7B | | Descriptor: | FAD-binding PCMH-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Haddad Momeni, M, Fredslund, F, Berrin, J.G, Abou Hachem, M, Welner, D.H. | | Deposit date: | 2020-04-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs.
Nat Commun, 12, 2021
|
|
8CEM
 
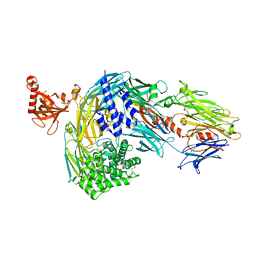 | | Structure of bovine native C3, re-refinement | | Descriptor: | Complement C3 alpha chain, Complement C3 beta chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Andersen, G.R, Fredslund, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of bovine complement component 3 reveals the basis for thioester function.
J Mol Biol, 361, 2006
|
|
6YWF
 
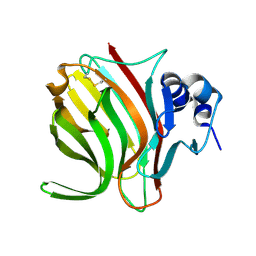 | |
4UNI
 
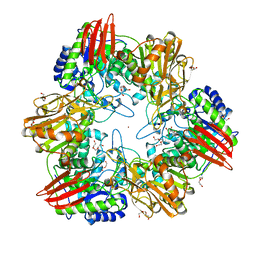 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
