1Y7Y
 
 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
2VRS
 
 | | Structure of avian reovirus sigmaC117-326, C2 crystal form | | Descriptor: | CHLORIDE ION, GLYCEROL, SIGMA-C CAPSID PROTEIN, ... | | Authors: | Guardado-Calvo, P, Fox, G.C, Llamas-Saiz, A.L, Benavente, J, van Raaij, M.J. | | Deposit date: | 2008-04-14 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre.
J. Gen. Virol., 90, 2009
|
|
2BSF
 
 | | Structure of the C-terminal receptor-binding domain of avian reovirus fibre sigmaC, Zn crystal form. | | Descriptor: | SIGMA C CAPSID PROTEIN, SULFATE ION, ZINC ION | | Authors: | Guardado Calvo, P, Fox, G.C, Hermo Parrado, X.L, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
J.Mol.Biol., 354, 2005
|
|
2VAK
 
 | | Crystal structure of the avian reovirus inner capsid protein sigmaA | | Descriptor: | SIGMA A, SULFATE ION | | Authors: | Guardado-Calvo, P, Llamas-Saiz, A.L, Fox, G.C, Hermo-Parrado, X.L, Vazquez-Iglesias, L, Martinez-Costas, J, Benavente, J, van Raaij, M.J. | | Deposit date: | 2007-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of the avian reovirus inner capsid protein sigmaA.
J.Virol., 82, 2008
|
|
2WT2
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with tri(N-acetyl-lactosamine) | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WT0
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with N-acetyl-lactosamine | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WSV
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lactose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2JJL
 
 | | Structure of avian reovirus sigmaC117-326, P321 crystal form | | Descriptor: | CHLORIDE ION, SIGMA-C CAPSID PROTEIN, SULFATE ION, ... | | Authors: | Guardado-Calvo, P, Fox, G.C, Llamas-Saiz, A.L, Benavente, J, van Raaij, M.J. | | Deposit date: | 2008-04-14 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre.
J. Gen. Virol., 90, 2009
|
|
2WT1
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lacto-N-neo-tetraose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
5ZMI
 
 | |
5JRM
 
 | | Crystal Structure of a Xylanase at 1.56 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, SULFATE ION | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
5JRN
 
 | | Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, methyl beta-D-xylopyranoside | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
2WSU
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | GLYCEROL, NITRATE ION, PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WST
 
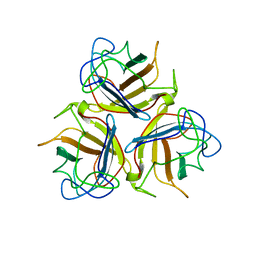 | | Head domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
6AC6
 
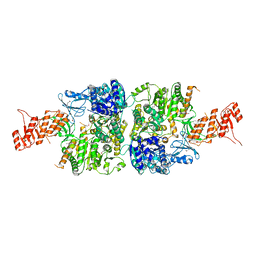 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AC8
 
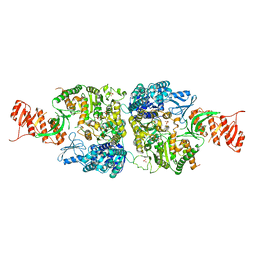 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
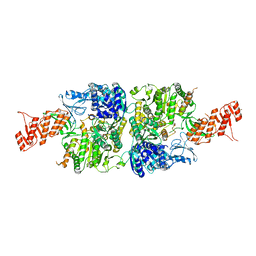 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACA
 
 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
2BT8
 
 | | Structure of the C-terminal receptor-binding domain of avian reovirus fibre sigmaC, space group P6322. | | Descriptor: | SIGMA C | | Authors: | Guardado Calvo, P, Fox, G.C, Hermo Parrado, X.L, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
J.Mol.Biol., 354, 2005
|
|
2BT7
 
 | | Structure of the C-terminal receptor-binding domain of avian reovirus fibre sigmaC, Cd crystal form | | Descriptor: | CADMIUM ION, SIGMA C, SULFATE ION | | Authors: | Guardado Calvo, P, Fox, G.C, Hermo Parrado, X.L, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
J.Mol.Biol., 354, 2005
|
|
2W37
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
2R9L
 
 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*DGP*DCP*DCP*DGP*DCP*DAP*DAP*DCP*DGP*DCP*DA)-3'), ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2007-09-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a NHEJ polymerase-mediated DNA synaptic complex
Science, 318, 2007
|
|
2Y77
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-2-ylmethoxy)-1,4,5- trihydroxy-2-(thiophen-2-ylmethyl)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[B]THIOPHEN-2-YL)METHOXY-1,4,5-TRIHYDROXY-2-(THIEN-2-YL)METHYLCYCLOHEX-2-EN-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2Y71
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-1,4,5-trihydroxy-3-((5-methylbenzo(b) thiophen-2-yl)methoxy)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2Y76
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-5-ylmethoxy)-2-(benzo(b) thiophen-5-ylmethyl)-1,4,5-trihydroxycyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[b]THIOPHEN-5-YL)METHOXY-2-(BENZO[b]THIOPHEN-5-YL)METHYL-1,4,5-TRIHYDROXYCYCLOHEX-2-ENE-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
