7OST
 
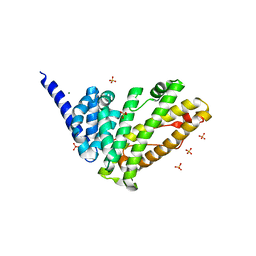 | | Rat HIP1R ANTH domain | | Descriptor: | Huntingtin-interacting protein 1-related, MAGNESIUM ION, SULFATE ION | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the N-terminal ANTH domain of rat HIP1R
To Be Published
|
|
6DEF
 
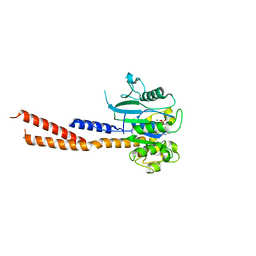 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
1W80
 
 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, bound to 2 peptides from Synaptojanin170 | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 2 ALPHA 2 SUBUNIT, BENZAMIDINE, CARBONATE ION, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving Nature of the Ap2 Alpha-Appendage Hub During Clathrin-Coated Vesicle Endocytosis.
Embo J., 23, 2004
|
|
2IV9
 
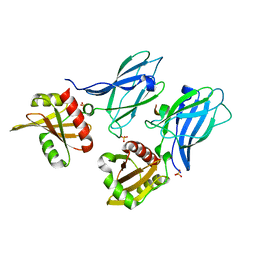 | |
2IV8
 
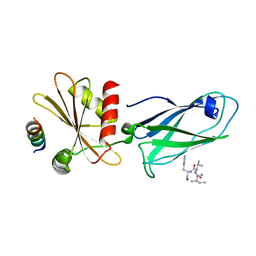 | |
2VJ0
 
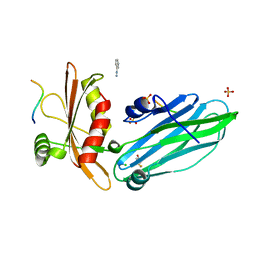 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, in complex with an FXDNF peptide from amphiphysin1 and a WVXF peptide from synaptojanin P170 | | Descriptor: | AMPHIPHYSIN, AP-2 COMPLEX SUBUNIT ALPHA-2, BENZAMIDINE, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solitary and Repetitive Binding Motifs for the Ap2 Complex {Alpha}-Appendage in Amphiphysin and Other Accessory Proteins.
J.Biol.Chem., 283, 2008
|
|
3ZVR
 
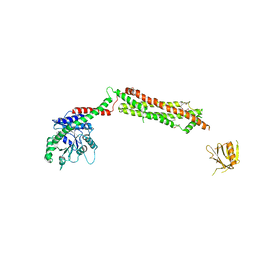 | |
1H0A
 
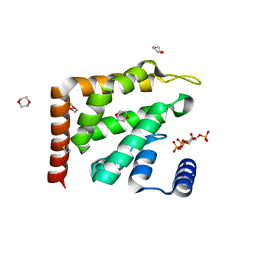 | | Epsin ENTH bound to Ins(1,4,5)P3 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, EPSIN | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2002-06-12 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Curvature of Clathrin-Coated Pits Driven by Epsin
Nature, 419, 2002
|
|
1HF8
 
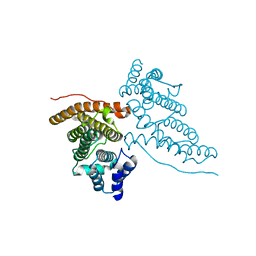 | |
1HFA
 
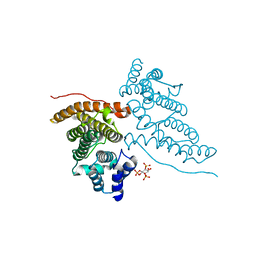 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, PI(4,5)P2 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-11-30 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
1HG5
 
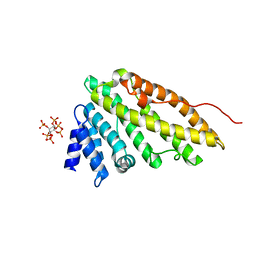 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, inositol(1,2,3,4,5,6)P6 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, INOSITOL HEXAKISPHOSPHATE | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-12-12 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
1HG2
 
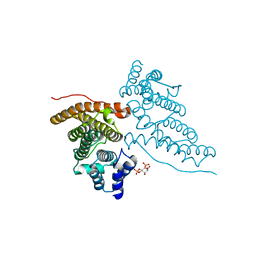 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, Inositol(4,5)P2 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, D-MYO-INOSITOL-4,5-BISPHOSPHATE | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-12-12 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
6DI7
 
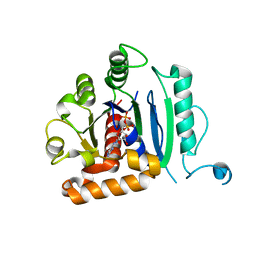 | | Vps1 GTPase-BSE fusion complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative sorting protein | | Authors: | Varlakhanova, N.V, Brady, T.M, Ford, M.G.J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DJQ
 
 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6Q0X
 
 | |
6VJF
 
 | | The P-Loop K to A mutation of C. therm Vps1 GTPase-BSE | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Putative sorting protein Vps1 | | Authors: | Tornabene, B.A, Varlakhanova, N.V, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural and functional characterization of the dominant negative P-loop lysine mutation in the dynamin superfamily protein Vps1.
Protein Sci., 29, 2020
|
|
