5FF2
 
 | | HydE from T. maritima in complex with (2R,4R)-TDA | | Descriptor: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
1KB5
 
 | | MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX | | Descriptor: | ANTIBODY DESIRE-1, KB5-C20 T-CELL ANTIGEN RECEPTOR | | Authors: | Housset, D, Mazza, G, Gregoire, C, Piras, C, Malissen, B, Fontecilla-Camps, J.C. | | Deposit date: | 1997-04-06 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain.
EMBO J., 16, 1997
|
|
3UQY
 
 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5FF0
 
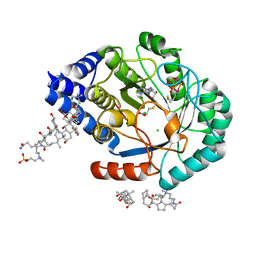 | | HydE from T. maritima in complex with S-adenosyl-L-cysteine and methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, Fe4-Se4 cluster, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
3USC
 
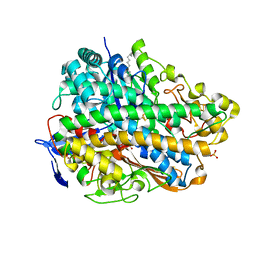 | | Crystal Structure of E. coli hydrogenase-1 in a ferricyanide-oxidized form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5F35
 
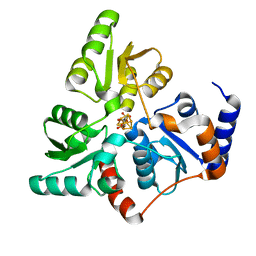 | | Structure of quinolinate synthase in complex with citrate | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F3D
 
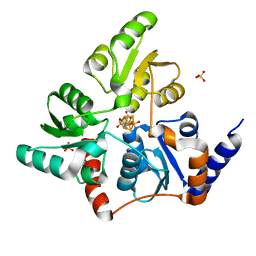 | | Structure of quinolinate synthase in complex with reaction intermediate W | | Descriptor: | 2-IMINO,3-CARBOXY,5-OXO,6-HYDROXY HEXANOIC ACID, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5F33
 
 | | Structure of quinolinate synthase in complex with phosphoglycolohydroxamate | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHOGLYCOLOHYDROXAMIC ACID, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
2Q60
 
 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
2FRV
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1997-06-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the [Nife] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands
J.Am.Chem.Soc., 118, 1996
|
|
2UZA
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Cavazza, C, Contreras-Martel, C, Fontecilla-Camps, J.C. | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate.
Structure, 14, 2006
|
|
6H8T
 
 | | Crystal structure of Papain modify by achiral Ru(II)complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Papain, ... | | Authors: | Cherrier, M.V, Amara, P, Talbi, B, Salmin, M, Fontecilla-Camps, J.C. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for unexpected selective tyrosine hydroxylations in an aerated achiral Ru-papain conjugate.
Metallomics, 10, 2018
|
|
2B3X
 
 | | Structure of an orthorhombic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, Iron-responsive element binding protein 1, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
2B3Y
 
 | | Structure of a monoclinic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
6HSE
 
 | | Structure of dithionite-reduced RsrR in spacegroup P2(1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Rrf2 family transcriptional regulator, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
6HSM
 
 | | Structure of partially reduced RsrR in space group P2(1)2(1)2(1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
6HSD
 
 | | Crystal structure of the oxidized form of the transcription regulator RsrR | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-09-30 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
1QDD
 
 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
1XIU
 
 | | Crystal structure of the agonist-bound ligand-binding domain of Biomphalaria glabrata RXR | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 1, RXR-like protein | | Authors: | De Groot, A, De Rosny, E, Juillan-Binard, C, Ferrer, J.-L, Laudet, V, Pebay-Peroula, E, Fontecilla-Camps, J.-C, Borel, F. | | Deposit date: | 2004-09-22 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Novel Tetrameric Complex of Agonist-bound Ligand-binding Domain of Biomphalaria glabrata Retinoid X Receptor.
J.Mol.Biol., 354, 2005
|
|
4R33
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan and S-adenosyl-L-homocysteine bound | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R34
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan, 5'-deoxyadenosine and methionine bound | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6I0R
 
 | | Structure of quinolinate synthase in complex with 5-mercaptopyridine-2,3-dicarboxylic acid | | Descriptor: | 5-mercaptopyridine-2,3-dicarboxylic acid, FE (III) ION, HYDROSULFURIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
1KEK
 
 | | Crystal Structure of the Free Radical Intermediate of Pyruvate:Ferredoxin Oxidoreductase | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Vernede, X, Guigliarelli, B, Charon, M.-H, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2001-11-16 | | Release date: | 2001-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase.
Science, 294, 2001
|
|
2C3Y
 
 | | CRYSTAL STRUCTURE OF THE RADICAL FORM OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-13 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
