1FRV
 
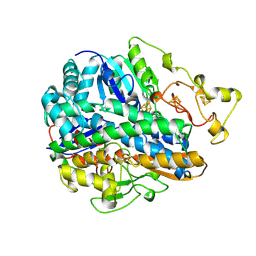 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | FE3-S4 CLUSTER, HYDRATED FE, HYDROGENASE, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas.
Nature, 373, 1995
|
|
1PK6
 
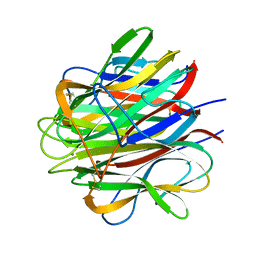 | | Globular Head of the Complement System Protein C1q | | Descriptor: | CALCIUM ION, Complement C1q subcomponent, A chain precursor, ... | | Authors: | Gaboriaud, C, Juanhuix, J, Gruez, A, Lacroix, M, Darnault, C, Pignol, D, Verger, D, Fontecilla-Camps, J.C, Arlaud, G.J. | | Deposit date: | 2003-06-05 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties.
J.Biol.Chem., 278, 2003
|
|
1ELV
 
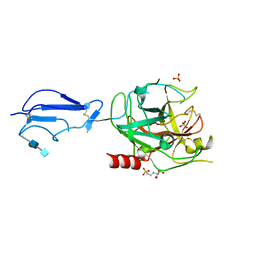 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1S COMPONENT, ... | | Authors: | Gaboriaud, C, Rossi, V, Bally, I, Arlaud, G, Fontecilla-Camps, J.-C. | | Deposit date: | 2000-03-14 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
EMBO J., 19, 2000
|
|
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
1NZI
 
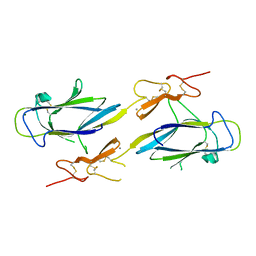 | | Crystal Structure of the CUB1-EGF Interaction Domain of Complement Protease C1s | | Descriptor: | CALCIUM ION, Complement C1s component, MAGNESIUM ION | | Authors: | Gregory, L.A, Thielens, N.M, Arlaud, G.J, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement
J.Biol.Chem., 278, 2003
|
|
1FAS
 
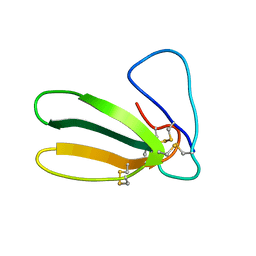 | | 1.9 ANGSTROM RESOLUTION STRUCTURE OF FASCICULIN 1, AN ANTI-ACETYLCHOLINESTERASE TOXIN FROM GREEN MAMBA SNAKE VENOM | | Descriptor: | FASCICULIN 1 | | Authors: | Le Du, M.H, Marchot, P, Bougis, P.E, Fontecilla-Camps, J.C. | | Deposit date: | 1992-08-07 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.9-A resolution structure of fasciculin 1, an anti-acetylcholinesterase toxin from green mamba snake venom.
J.Biol.Chem., 267, 1992
|
|
1OAO
 
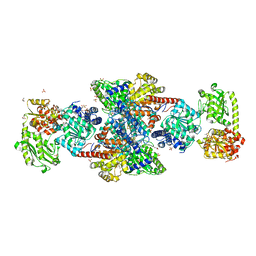 | | NiZn[Fe4S4] and NiNi[Fe4S4] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase | | Descriptor: | ACETATE ION, BICARBONATE ION, CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Darnault, C, Volbeda, A, Kim, E.J, Legrand, P, Vernede, X, Lindahl, P.A, Fontecilla-Camps, J.C. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Alpha Subunits of Acetyl-Coa Synthase/Carbon Monoxide Dehydrogenase
Nat.Struct.Biol., 10, 2003
|
|
4JXC
 
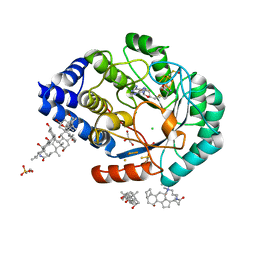 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JY9
 
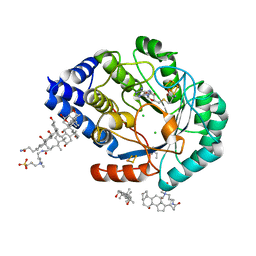 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYF
 
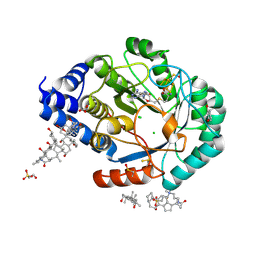 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYE
 
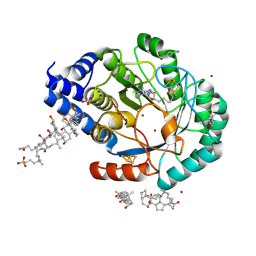 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | BROMIDE ION, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYD
 
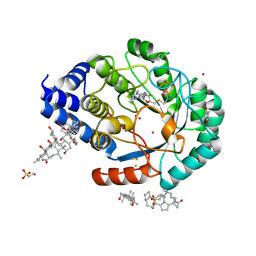 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, BROMIDE ION, CHAPSO, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JY8
 
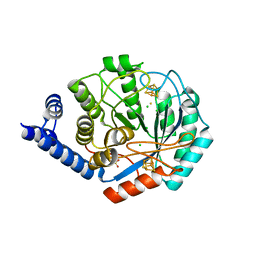 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHLORIDE ION, FEFE-HYDROGENASE MATURASE, HYDROSULFURIC ACID, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DDI
 
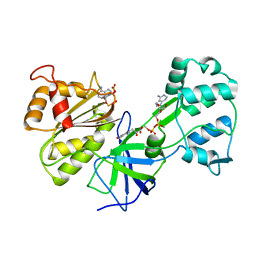 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDG
 
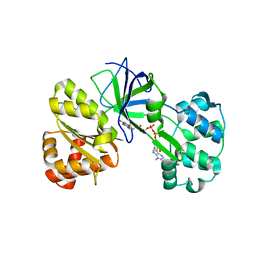 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1E08
 
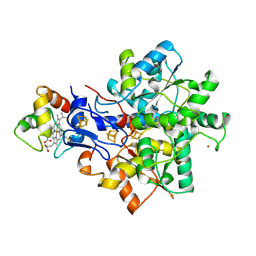 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
1FDS
 
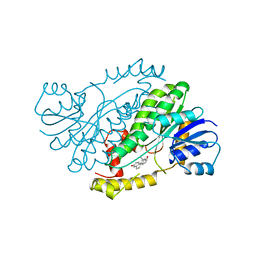 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH 17-BETA-ESTRADIOL | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
1FDT
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
1ETH
 
 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
1EYX
 
 | | CRYSTAL STRUCTURE OF R-PHYCOERYTHRIN AT 2.2 ANGSTROMS | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOUROBILIN, ... | | Authors: | Contreras-Martel, C, Legrand, P, Piras, C, Vernede, X, Martinez-Oyanedel, J, Bunster, M, Fontecilla-Camps, J.C. | | Deposit date: | 2000-05-09 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallization and 2.2 A resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FON
 
 | | CRYSTAL STRUCTURE OF BOVINE PROCARBOXYPEPTIDASE A-S6 SUBUNIT III, A HIGHLY STRUCTURED TRUNCATED ZYMOGEN E | | Descriptor: | PROCARBOXYPEPTIDASE A-S6 | | Authors: | Pignol, D.C, Gaboriaud, T, Michon, B, Kerfelec, B, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of bovine procarboxypeptidase A-S6 subunit III, a highly structured truncated zymogen E.
EMBO J., 13, 1994
|
|
