8A9Q
 
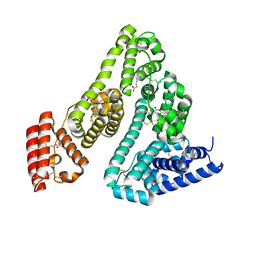 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
6FHE
 
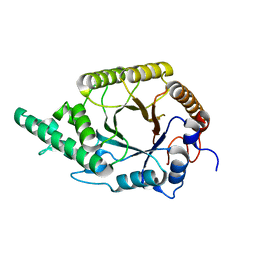 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Synthetic construct | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
6FHF
 
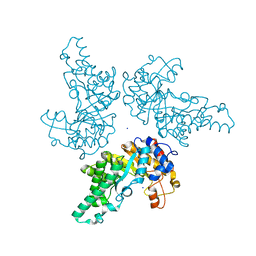 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Design, SODIUM ION | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
