1ASJ
 
 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR9
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR6
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
433D
 
 | |
1TME
 
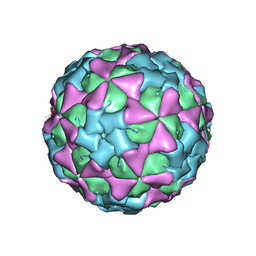 | | THREE-DIMENSIONAL STRUCTURE OF THEILER VIRUS | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1992-01-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of Theiler virus.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
5DOE
 
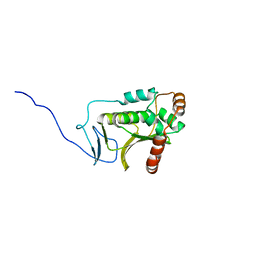 | | Crystal structure of the Human Cytomegalovirus UL53 (residues 72-292) | | Descriptor: | Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
5DOB
 
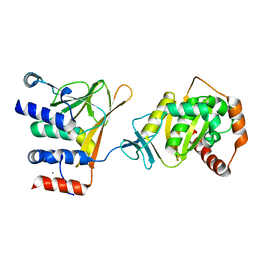 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
5DOC
 
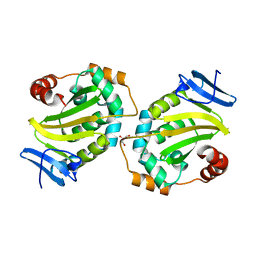 | | Crystal structure of the Human Cytomegalovirus UL53 subunit of the NEC | | Descriptor: | GLYCEROL, Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
5KTZ
 
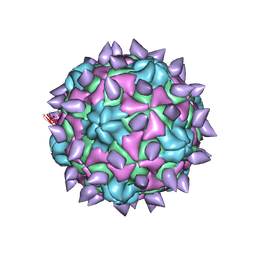 | | expanded poliovirus in complex with VHH 12B | | Descriptor: | Genome polyprotein, VHH 12B, VP1, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KU0
 
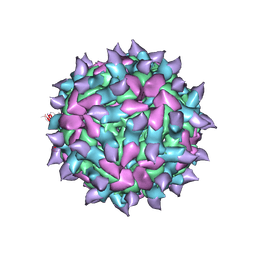 | | expanded poliovirus in complex with VHH 17B | | Descriptor: | VHH 17B, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
2IJF
 
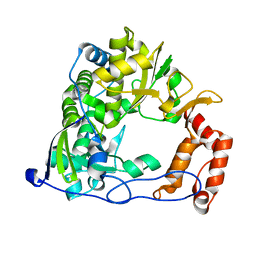 | |
2IJD
 
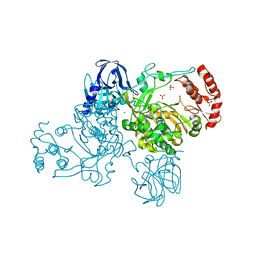 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
2O0I
 
 | | crystal structure of the R185A mutant of the N-terminal domain of the Group B Streptococcus Alpha C protein | | Descriptor: | C protein alpha-antigen | | Authors: | Hogle, J.M, Filman, D.J, Baron, M.J, Madoff, L.C, Iglesias, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of a glycosaminoglycan binding region of the alpha C protein that mediates entry of group B streptococci into host cells.
J.Biol.Chem., 282, 2007
|
|
5KU2
 
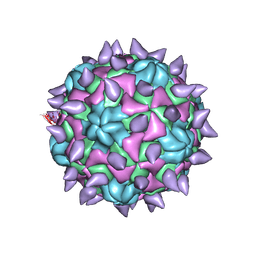 | | expanded poliovirus in complex with VHH 7A | | Descriptor: | VHH 7A, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KWL
 
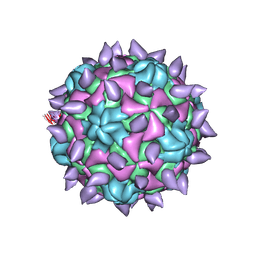 | | expanded poliovirus in complex with VHH 10E | | Descriptor: | VHH 10E, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-18 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
6P9W
 
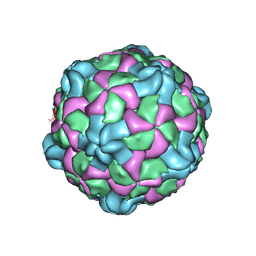 | | Poliovirus (Type 1 Mahoney), receptor catalysed 135S particle map | | Descriptor: | VP1, VP2, VP3 | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
6P9O
 
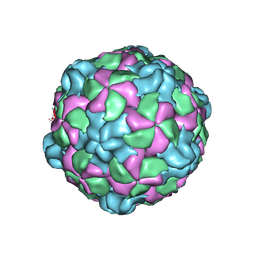 | |
6PSZ
 
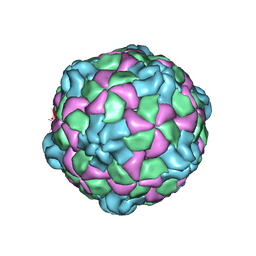 | | Poliovirus (Type 1 Mahoney), heat-catalysed 135S particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-07-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
6Q0B
 
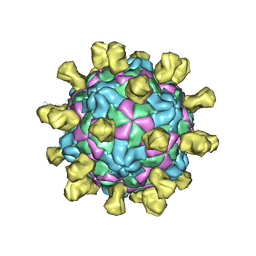 | | Poliovirus (Type 1 Mahoney), receptor-catalysed 135S particle incubated with anti-VP1 mAb at RT for 1 hr | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
1XOK
 
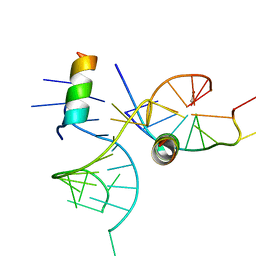 | | crystal structure of alfalfa mosaic virus RNA 3'UTR in complex with coat protein N terminal peptide | | Descriptor: | BROMIDE ION, Coat protein, alfalfa mosaic virus RNA 3' UTR | | Authors: | Guogas, L.M, Filman, D.J, Hogle, J.M, Gehrke, L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cofolding organizes alfalfa mosaic virus RNA and coat protein for replication.
Science, 306, 2004
|
|
1YYP
 
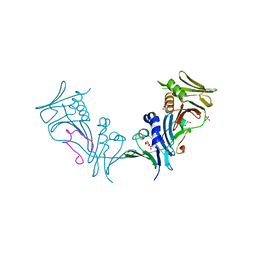 | | Crystal structure of cytomegalovirus UL44 bound to C-terminal peptide from CMV UL54 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Appleton, B.A, Brooks, J, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2005-02-25 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.
J.Biol.Chem., 281, 2006
|
|
1XYR
 
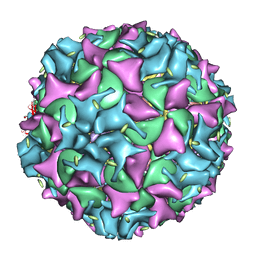 | | Poliovirus 135S cell entry intermediate | | Descriptor: | Genome polyprotein, Coat protein VP1, Coat protein VP2, ... | | Authors: | Bubeck, D, Filman, D.J, Cheng, N, Steven, A.C, Hogle, J.M, Belnap, D.M. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
J.Virol., 79, 2005
|
|
1YWM
 
 | | Crystal structure of the N-terminal domain of group B Streptococcus alpha C protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C protein alpha-antigen, GLYCEROL | | Authors: | Auperin, T.C, Bolduc, G.R, Baron, M.J, Heroux, A, Filman, D.J, Madoff, L.C, Hogle, J.M. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein.
J.Biol.Chem., 280, 2005
|
|
1EV1
 
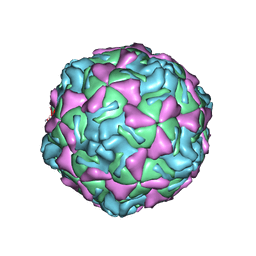 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1T6L
 
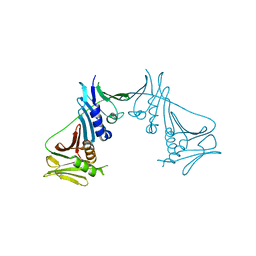 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
