1Y1H
 
 | | human formylglycine generating enzyme, oxidised Cys refined as hydroperoxide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-alpha-formyglycine-generating enzyme, HYDROGEN PEROXIDE, ... | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Y1J
 
 | | human formylglycine generating enzyme, sulfonic acid/desulfurated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-alpha-formyglycine-generating enzyme, ... | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Y4J
 
 | | Crystal structure of the paralogue of the human formylglycine generating enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dickmanns, A, Rudolph, M.G, Ficner, R. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal Structure of Human pFGE, the Paralog of the C{alpha}-formylglycine-generating Enzyme
J.Biol.Chem., 280, 2005
|
|
5LTK
 
 | |
6H42
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
6H45
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
3JQY
 
 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | Authors: | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
5LZA
 
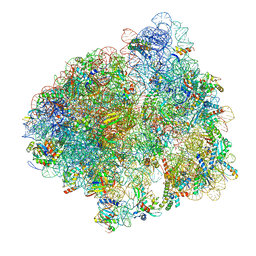 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZF
 
 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZD
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZC
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5D0U
 
 | |
4XRI
 
 | | Crystal Structure of Importin Beta in an Ammonium Sulfate Condition | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION | | Authors: | Tauchert, M.J, Neumann, P, Ficner, R, Dickmanns, A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impact of the crystallization condition on importin-beta conformation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1V0E
 
 | | Endosialidase of Bacteriophage K1F | | Descriptor: | ENDO-ALPHA-SIALIDASE, PHOSPHATE ION | | Authors: | Stummeyer, K, Dickmanns, A, Muehlenhoff, M, Gerady-Schahn, R, Ficner, R. | | Deposit date: | 2004-03-28 | | Release date: | 2004-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Polysialic Acid-Degrading Endosialidase of Bacteriophage K1F
Nat.Struct.Mol.Biol., 12, 2005
|
|
1V0F
 
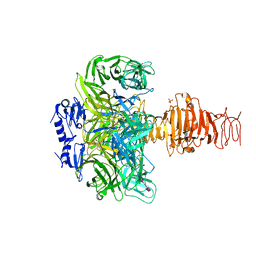 | | Endosialidase of Bacteriophage K1F in complex with oligomeric alpha-2,8-sialic acid | | Descriptor: | ENDO-ALPHA-SIALIDASE, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid, ... | | Authors: | Stummeyer, K, Dickmanns, A, Muehlenhoff, M, Gerady-Schahn, R, Ficner, R. | | Deposit date: | 2004-03-28 | | Release date: | 2004-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Polysialic Acid-Degrading Endosialidase of Bacteriophage K1F
Nat.Struct.Mol.Biol., 12, 2005
|
|
5AFI
 
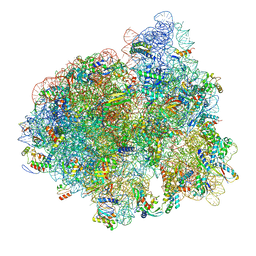 | | 2.9A Structure of E. coli ribosome-EF-TU complex by cs-corrected cryo-EM | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Konevega, A.L, Bock, L.V, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the E. coli ribosome-EF-Tu complex at <3 angstrom resolution by Cs-corrected cryo-EM.
Nature, 520, 2015
|
|
6FAC
 
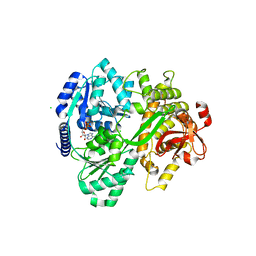 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
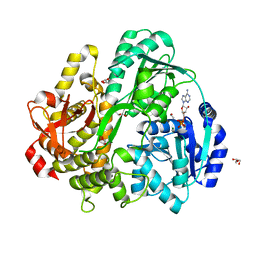 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4H6B
 
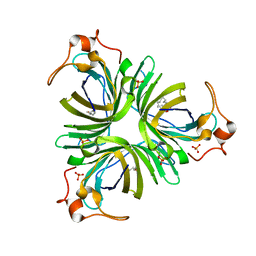 | | Structural basis for allene oxide cyclization in moss | | Descriptor: | (9Z)-11-{(2R,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, (9Z)-11-{(2S,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, Allene oxide cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
5J8N
 
 | |
3FGR
 
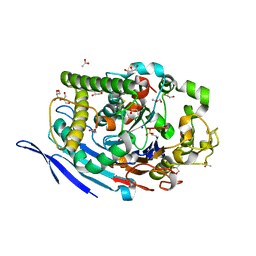 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGT
 
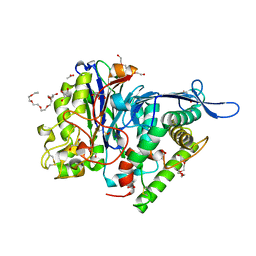 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
4XRK
 
 | |
