1W4G
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state folding transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4H
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4J
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family.
J. Mol. Biol., 353, 2005
|
|
1W4E
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4K
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4I
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1YPC
 
 | |
1YPB
 
 | |
1YPA
 
 | |
2C6A
 
 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
1ZT4
 
 | | The crystal structure of human CD1d with and without alpha-Galactosylceramide | | Descriptor: | Beta-2-microglobulin, N-{(1S,2R,3S)-1-[(ALPHA-D-GALACTOPYRANOSYLOXY)METHYL]-2,3-DIHYDROXYHEPTADECYL}HEXACOSANAMIDE, T-cell surface glycoprotein CD1d | | Authors: | Koch, M, Stronge, V.S, Shepherd, D, Gadola, S.D, Mathew, B, Ritter, G, Fersht, A.R, Besra, G.S, Schmidt, R.R, Jones, E.Y, Cerundolo, V, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of human CD1d with and without alpha-galactosylceramide
Nat.Immunol., 6, 2005
|
|
1ZTR
 
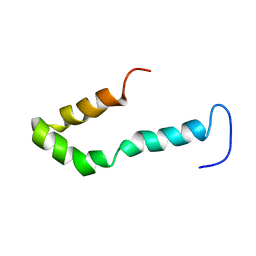 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
2C6B
 
 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
1K5R
 
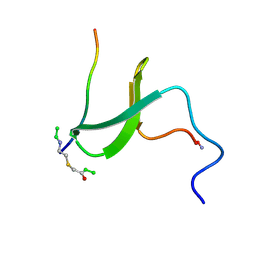 | | YAP65 WW domain S24-Amino-Ethylsulfanyl-Acetic Acid mutant | | Descriptor: | 65 KDA YES-ASSOCIATED PROTEIN, Fragment of WBP-1 | | Authors: | Ferguson, N, Pires, J.R, Toepert, F, Johnson, C.M, Pan, Y.P, Volkmer-Engert, R, Schneider-Mergener, J, Daggett, V, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2001-10-12 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Using flexible loop mimetics to extend phi-value analysis to secondary structure interactions.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2J1X
 
 | |
2J1W
 
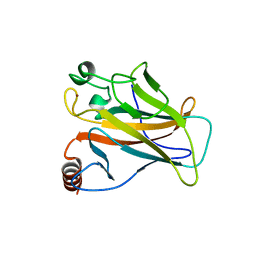 | |
2J1Z
 
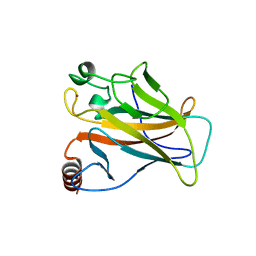 | |
2J21
 
 | |
2J1Y
 
 | |
2J20
 
 | |
1CIR
 
 | | COMPLEX OF TWO FRAGMENTS OF CI2 [(1-40)(DOT)(41-64)] | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Davis, B.J, Fersht, A.R. | | Deposit date: | 1995-10-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards the complete structural characterization of a protein folding pathway: the structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism
Structure Fold.Des., 1, 1996
|
|
1CIQ
 
 | | COMPLEX OF TWO FRAGMENTS OF CI2, RESIDUES 1-40 AND 41-64 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Buckle, A.M, Fersht, A.R. | | Deposit date: | 1995-10-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Towards the complete structural characterization of a protein folding pathway: the structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism
Structure Fold.Des., 1, 1996
|
|
1B27
 
 | |
1B2S
 
 | |
