4UYP
 
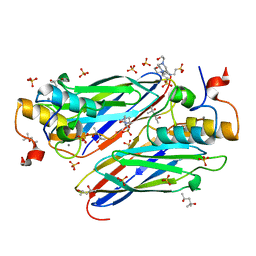 | | High resolution structure of the third cohesin ScaC in complex with the ScaB dockerin with a mutation in the N-terminal helix (IN to SI) from Acetivibrio cellulolyticus displaying a type I interaction. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Cameron, K, Alves, V.D, Bule, P, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
J. Biol. Chem., 290, 2015
|
|
4V1L
 
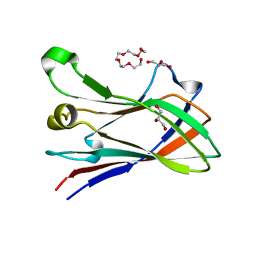 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UZN
 
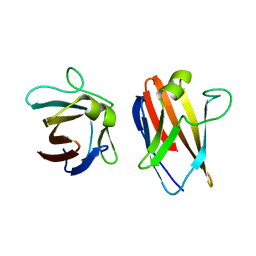 | | The native structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B) | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4V1I
 
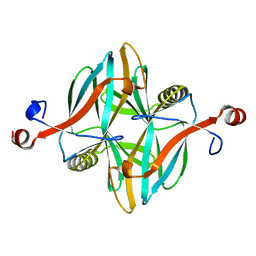 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 at medium resolution | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8PN6
 
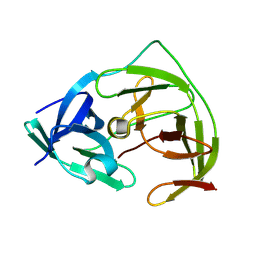 | | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus | | Descriptor: | Genome polyprotein, Serine protease subunit NS2B | | Authors: | Ni, X, Fairhead, M, Balcomb, B.H, Aschenbrenner, J.C, Ferreira, L.M, Godoy, A.S, Lithgo, R.M, MacLean, E.M, Marples, P.G, Thompson, W, Tomlinson, C.W.E, Szommer, T, Wild, C, Wright, N.D, Koekemoer, L, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus
To Be Published
|
|
1OHZ
 
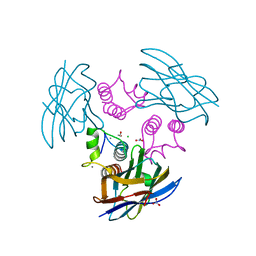 | | Cohesin-Dockerin complex from the cellulosome of Clostridium thermocellum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4V18
 
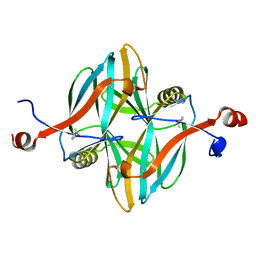 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V2X
 
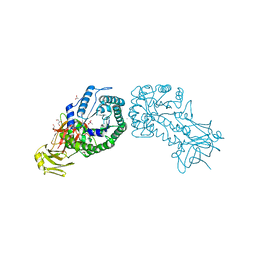 | | High resolution structure of the full length tri-modular endo-beta-1, 4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
2YPJ
 
 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
4UZ8
 
 | | The SeMet structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B), SULFATE ION | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4V1B
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 collected at the Zn edge | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V17
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V1K
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | 2-HYDROXY BUTANE-1,4-DIOL, CALCIUM ION, CARBOHYDRATE BINDING MODULE, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UMS
 
 | | The crystal structure of the seventh ScaB type I cohesin from Pseudobacteroides cellulosolvens | | Descriptor: | CELLULOSOMAL ANCHORING SCAFFOLDIN B | | Authors: | Cameron, K, Alves, V.D, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Combined Crystal Structure of a Type-I Cohesin, Mutation and Affinity-Binding Studies Reveal Structural Determinants of Cohesin-Dockerin Specificity
J.Biol.Chem., 290, 2015
|
|
1UR2
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
2H5Y
 
 | | Crystallographic structure of the Molybdate-Binding Protein of Xanthomonas citri at 1.7 Ang resolution bound to molybdate | | Descriptor: | MOLYBDATE ION, Molybdate-binding periplasmic protein, SULFATE ION | | Authors: | Balan, A, Santacruz, C.P, Ferreira, L.C.S, Barbosa, J.A.R.G. | | Deposit date: | 2006-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallization, data collection and phasing of the molybdate-binding protein of the phytopathogen Xanthomonas axonopodis pv. citri
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1UQZ
 
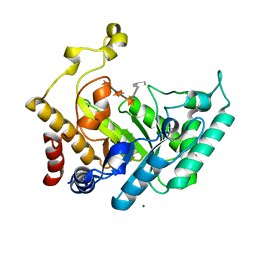 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ENDOXYLANASE, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1GKL
 
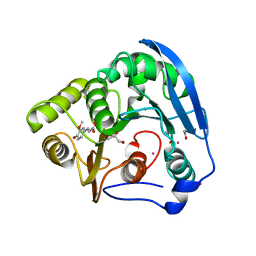 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ACETATE ION, CADMIUM ION, ... | | Authors: | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition.
Structure, 9, 2001
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6X
 
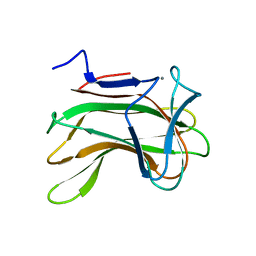 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
2C4X
 
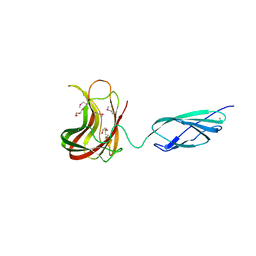 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
1UR1
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
