6J2Y
 
 | |
6K4Y
 
 | | CryoEM structure of sigma appropriation complex | | 分子名称: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2019-05-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
7D1O
 
 | | Crystal structure of SARS-Cov-2 main protease with narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Fu, L.F, Feng, Y, Qi, J.X. | | 登録日 | 2020-09-15 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis for the inhibition of the SARS-CoV-2 main protease by the anti-HCV drug narlaprevir.
Signal Transduct Target Ther, 6, 2021
|
|
7C78
 
 | |
7ECV
 
 | | The Csy-AcrIF14 complex | | 分子名称: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECW
 
 | | The Csy-AcrIF14-dsDNA complex | | 分子名称: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7W7H
 
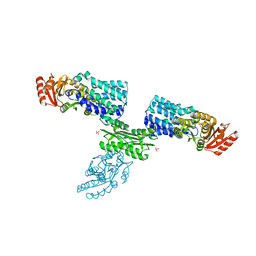 | | S Suis FakA-FakB2 complex structure | | 分子名称: | OLEIC ACID, Predicted kinase related to dihydroxyacetone kinase, SULFATE ION, ... | | 著者 | Shi, Y, Zang, N, Lou, N, Xu, Y, Sun, J, Huang, M, Zhang, H, Lu, H, Zhou, C, Feng, Y. | | 登録日 | 2021-12-04 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and mechanism for streptococcal fatty acid kinase (Fak) system dedicated to host fatty acid scavenging.
Sci Adv, 8, 2022
|
|
7XYA
 
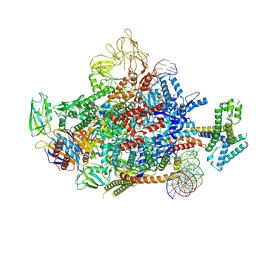 | | The cryo-EM structure of an AlpA-loading complex | | 分子名称: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Wen, A, Feng, Y. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XYB
 
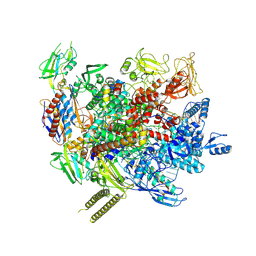 | | The cryo-EM structure of an AlpA-loaded complex | | 分子名称: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Wen, A, Feng, Y. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
6KG3
 
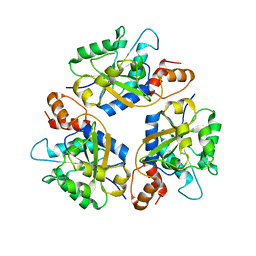 | |
4NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: D58P OXIDIZED | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | 登録日 | 1996-12-13 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57D OXIDIZED | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | 登録日 | 1996-12-16 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4PMD
 
 | |
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2021-03-24 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
7ELM
 
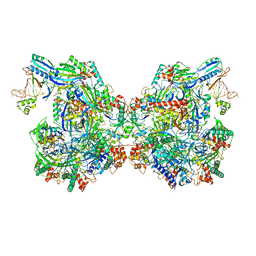 | | Structure of Csy-AcrIF24 | | 分子名称: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L, Feng, Y. | | 登録日 | 2021-04-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7EQG
 
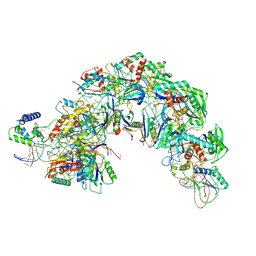 | | Structure of Csy-AcrIF5 | | 分子名称: | AcrIF5, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L, Feng, Y. | | 登録日 | 2021-05-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
6J92
 
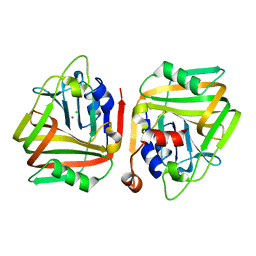 | |
7CG8
 
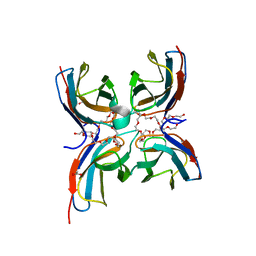 | | Structure of the sensor domain (short construct) of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, Anti-sigma factor RsgI, ... | | 著者 | Dong, S, Feng, Y. | | 登録日 | 2020-06-30 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the sensor domain of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens
To Be Published
|
|
7D5G
 
 | |
7WDH
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NZS
 
 | | Dextranase AoDex KQ11 | | 分子名称: | Dextranase | | 著者 | Ren, W, Yan, W, Gu, L, Feng, Y, Dong, D, Wang, S, Wang, C, Lyu, M. | | 登録日 | 2019-02-14 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
To Be Published
|
|
7Y0S
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-tyrosyl-L-tyrosine and hydroxylamine | | 分子名称: | Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, I7X-TYR-TYR, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2022-06-06 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-tyrosyl-L-tyrosine and hydroxylamine
To Be Published
|
|
7Y0R
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, 4-METHYLANILINE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2022-06-06 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine
To Be Published
|
|
7Y0Q
 
 | |
