1AGZ
 
 | |
1AGK
 
 | |
1AGO
 
 | |
4CSU
 
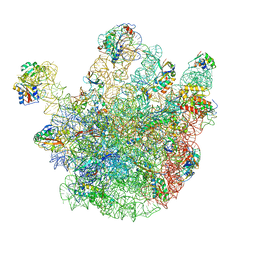 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
1AGH
 
 | |
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
2YKR
 
 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
