2LWF
 
 | | Structure of N-terminal domain of a plant Grx | | 分子名称: | Monothiol glutaredoxin-S16, chloroplastic | | 著者 | Feng, Y. | | 登録日 | 2012-07-28 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LEO
 
 | |
2K6B
 
 | |
2JTM
 
 | | Solution structure of Sso6901 from Sulfolobus solfataricus P2 | | 分子名称: | Putative uncharacterized protein | | 著者 | Feng, Y, Guo, L, Huang, L, Wang, J. | | 登録日 | 2007-08-03 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea
Nucleic Acids Res., 36, 2008
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | 分子名称: | Protein male-specific lethal-2, ZINC ION | | 著者 | Feng, Y, Ye, K, Zheng, S, Wang, J. | | 登録日 | 2012-06-09 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
2MMP
 
 | | Solution structure of a ribosomal protein | | 分子名称: | Uncharacterized protein | | 著者 | Feng, Y. | | 登録日 | 2014-03-17 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of archaea-specific ribosomal protein L46a reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 450, 2014
|
|
2KJG
 
 | |
6U1T
 
 | | Crystal structure of anti-Nipah virus (NiV) F 5B3 antibody Fab fragment | | 分子名称: | CHLORIDE ION, antigen-binding (Fab) fragment, heavy chain, ... | | 著者 | Dang, H.V, Chan, Y.P, Park, Y.J, Snijder, J, Da Silva, S.C, Vu, B, Yan, L, Feng, Y.R, Rockx, B, Geisbert, T, Mire, C, Mire, C.E, BBroder, C.C, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-16 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6LBP
 
 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | 分子名称: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | 著者 | Yi, Z, Cao, X, Han, F, Feng, Y. | | 登録日 | 2019-11-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.065 Å) | | 主引用文献 | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
6M6B
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
1FVX
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57N OXIDIZED | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | 登録日 | 1996-12-12 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4W4V
 
 | | JNK2/3 in complex with 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | 分子名称: | 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-Jun N-terminal kinase 3 | | 著者 | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | 登録日 | 2014-08-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4Y
 
 | | JNK2/3 in complex with 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | 分子名称: | 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | 著者 | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | 登録日 | 2014-08-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4W
 
 | | JNK2/3 in complex with N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide | | 分子名称: | N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide, c-Jun N-terminal kinase 3 | | 著者 | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | 登録日 | 2014-08-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
6DVD
 
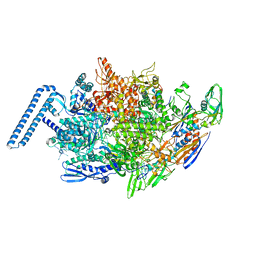 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) with 6 nt spacer and bromine labelled in position "-11 | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | 登録日 | 2018-06-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.899 Å) | | 主引用文献 | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVB
 
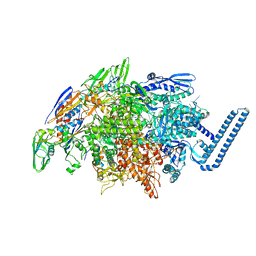 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 5nt spacer | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | 登録日 | 2018-06-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DV9
 
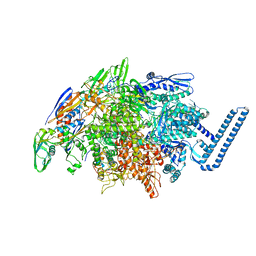 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 4nt spacer | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | 登録日 | 2018-06-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVE
 
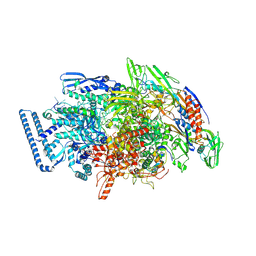 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF selenomethionine-labelled sigma factor L) with 6 nt spacer | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*GP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | 登録日 | 2018-06-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2019-02-27 | | 実験手法 | X-RAY DIFFRACTION (3.812 Å) | | 主引用文献 | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
4TXT
 
 | |
1APC
 
 | |
4W4X
 
 | | JNK2/3 in complex with 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | 分子名称: | 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | 著者 | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | 登録日 | 2014-08-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
6TYS
 
 | | A potent cross-neutralizing antibody targeting the fusion glycoprotein inhibits Nipah virus and Hendra virus infection | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5B3 antibody heavy chain, ... | | 著者 | Dang, H.V, Chan, Y.P, Park, Y.J, Snijder, J, Da Silva, S.C, Vu, B, Yan, L, Feng, Y.R, Rockx, B, Geisbert, T, Mire, C.E, Broder, C.B, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-08-09 | | 公開日 | 2019-10-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8YSA
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H102 | | 分子名称: | 3C-like proteinase nsp5, BOC-TBG-PHE-ELL | | 著者 | Zheng, W.Y, Fu, L.F, Feng, Y, Han, P, Qi, J.X. | | 登録日 | 2024-03-22 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery, Biological Activity, and Structural Mechanism of a Potent Inhibitor of SARS-CoV-2 Main Protease
To Be Published
|
|
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6C
 
 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
