6A9Y
 
 | | The crystal structure of Mu homology domain of SGIP1 | | 分子名称: | SH3-containing GRB2-like protein 3-interacting protein 1 | | 著者 | Feng, Y, Liu, X. | | 登録日 | 2018-07-16 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | SGIP1 dimerizes via intermolecular disulfide bond in mu HD domain during cellular endocytosis.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5GL5
 
 | |
4JND
 
 | | Structure of a C.elegans sex determining protein | | 分子名称: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | 著者 | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | 登録日 | 2013-03-15 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.652 Å) | | 主引用文献 | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
5X39
 
 | |
5X34
 
 | |
7Y67
 
 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | 分子名称: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y65
 
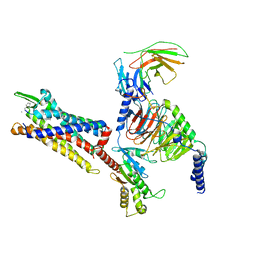 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | 分子名称: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
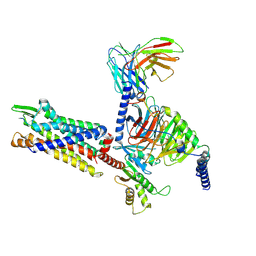 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | 分子名称: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
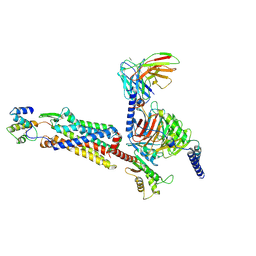 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | 分子名称: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
5X38
 
 | |
5X35
 
 | |
5X36
 
 | |
5X37
 
 | |
5X3C
 
 | |
5XVM
 
 | |
7XSP
 
 | | Structure of gRAMP-target RNA | | 分子名称: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | 著者 | Feng, Y, Zhang, L.X. | | 登録日 | 2022-05-15 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSR
 
 | | Structure of Craspase-target RNA | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | 著者 | Feng, Y, Zhang, L. | | 登録日 | 2022-05-15 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
 | | Structure of the Craspase | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | 著者 | Feng, Y, Zhang, L. | | 登録日 | 2022-05-15 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XT4
 
 | | Structure of Craspase-NTR | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | 著者 | Feng, Y, Zhang, L. | | 登録日 | 2022-05-16 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSS
 
 | | Structure of Craspase-CTR | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | 著者 | Feng, Y, Zang, L.X. | | 登録日 | 2022-05-15 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSO
 
 | |
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | 分子名称: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Feng, Y, Zhang, L.X. | | 登録日 | 2022-07-14 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
5HWH
 
 | |
8H2X
 
 | | Structure of Acb2 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | 著者 | Feng, Y, Cao, X.L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
 | | Structure of Acb2 complexed with c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | 著者 | Feng, Y, Cao, X.L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
