7GZL
 
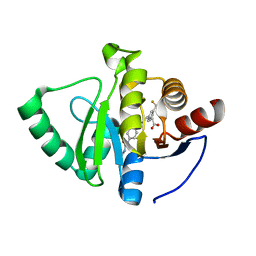 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008304-001 | | Descriptor: | (3R)-3-[4-(cyclopropylcarbamamido)benzamido]-3-[3-(difluoromethyl)phenyl]propanoic acid, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZI
 
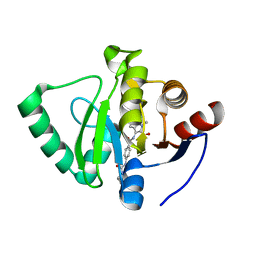 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000753-001 | | Descriptor: | 3-[4-(cyclopropylcarbamamido)benzamido]-1-methyl-1H-pyrrolo[2,3-b]pyridine-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZF
 
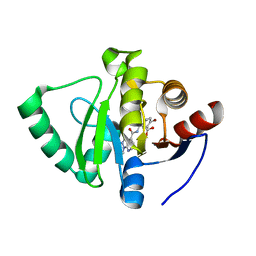 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000605-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1-methyl-1H-pyrazolo[3,4-b]pyridine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ4
 
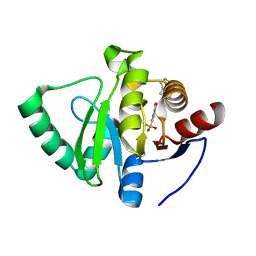 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000018-001 | | Descriptor: | 5-[(2-fluorophenyl)sulfamoyl]-2-methyl-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZH
 
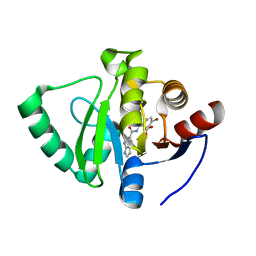 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000670-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[2,3-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZG
 
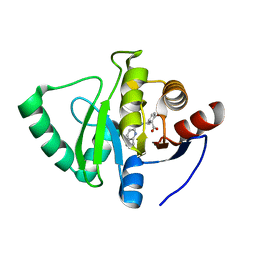 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000620-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[3,2-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ7
 
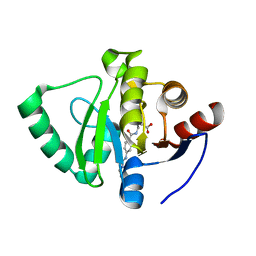 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000131-001 | | Descriptor: | 3-{[4-(cyclopropylcarbamamido)benzamido]methyl}-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ1
 
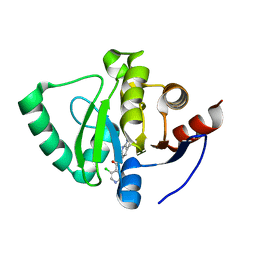 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000061-001 | | Descriptor: | 3-chloro-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)pyridine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZE
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000601-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-{[5-(dimethylamino)pyridine-2-carbonyl]amino}propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GQH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1509711879 | | Descriptor: | 6-bromo-7-hydroxy-2,2-dimethyl-2H,4H-1,3-benzodioxin-4-one, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GO5
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198275935 | | Descriptor: | 6,8-bis(fluoranyl)chromene, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z220996120 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(1S)-1-(pyridin-3-yl)ethyl]cyclopropanecarboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPE
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z760031264 | | Descriptor: | 4-(1,2,4-oxadiazol-5-yl)aniline, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPK
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z905434478 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with NCL-00024661 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQ1
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z166605460 | | Descriptor: | Protease 3C, thiophene-3-carboxylic acid | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQ9
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z281773378 | | Descriptor: | 4-(4-methoxyphenyl)-1,2,3,6-tetrahydropyridine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQG
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z228589380 | | Descriptor: | 3-[(pyrimidin-2-yl)amino]benzoic acid, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GNV
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z104474228 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GO0
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1137725943 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(1H-indol-4-yl)methyl]ethanamine, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GO2
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1182328459 | | Descriptor: | (4S)-[1,2,4]triazolo[4,3-a]pyrazine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GO9
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1266933824 | | Descriptor: | (1H-pyrazol-4-yl)(pyrrolidin-1-yl)methanone, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOB
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1295863442 | | Descriptor: | 3-fluoro-5-methylbenzene-1-sulfonamide, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z137811222 | | Descriptor: | 1-{4-[(1E)-3-phenylprop-1-en-1-yl]piperazin-1-yl}ethan-1-one, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOG
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1398461996 | | Descriptor: | (1R)-1-(3-benzyl-1,2,4-oxadiazol-5-yl)ethan-1-amine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
