4RI2
 
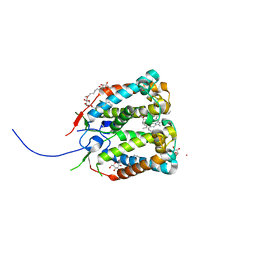 | | Crystal structure of the photoprotective protein PsbS from spinach | | Descriptor: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RI3
 
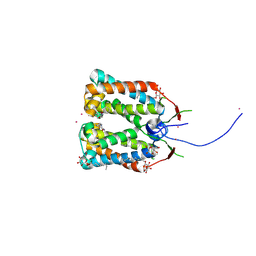 | | Crystal structure of DCCD-modified PsbS from spinach | | Descriptor: | DICYCLOHEXYLUREA, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5IE2
 
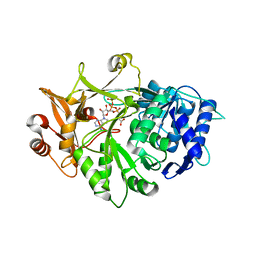 | | Crystal structure of a plant enzyme | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fan, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
5IE3
 
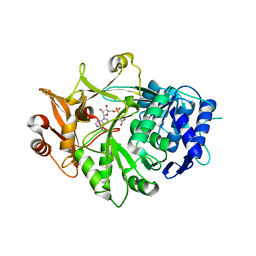 | | Crystal structure of a plant enzyme | | Descriptor: | ADENOSINE MONOPHOSPHATE, OXALIC ACID, Oxalate--CoA ligase | | Authors: | Fan, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
6HK5
 
 | | X-ray structure of a truncated mutant of the metallochaperone CooJ with a high-affinity nickel-binding site | | Descriptor: | 3,3',3''-phosphoryltripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Alfano, M, Perard, J, Basset, C, Carpentier, P, Zambelli, B, Timm, J, Crouzy, S, Ciurli, S, Cavazza, C. | | Deposit date: | 2018-09-05 | | Release date: | 2019-03-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | The carbon monoxide dehydrogenase accessory protein CooJ is a histidine-rich multidomain dimer containing an unexpected Ni(II)-binding site.
J.Biol.Chem., 294, 2019
|
|
6C5W
 
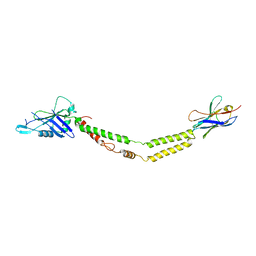 | | Crystal structure of the mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, calcium uniporter, nanobody | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-11 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.10010242 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6C5R
 
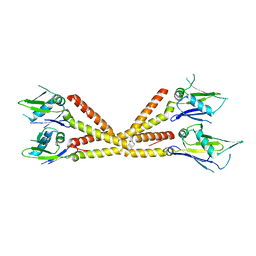 | | Crystal structure of the soluble domain of the mitochondrial calcium uniporter | | Descriptor: | calcium uniporter | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-16 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09608316 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
8YBR
 
 | | Choline transporter BetT | | Descriptor: | BCCT family transporter | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
8YBQ
 
 | | Choline transporter BetT - CHT bound | | Descriptor: | BCCT family transporter, CHOLINE ION | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
6WDO
 
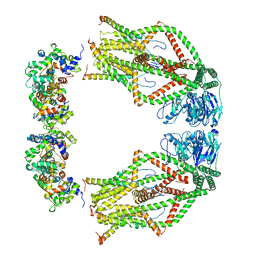 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
6WDN
 
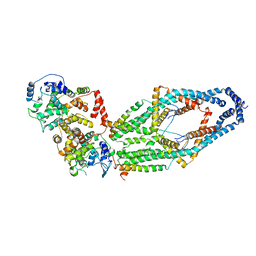 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in low Ca2+ | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
8FHN
 
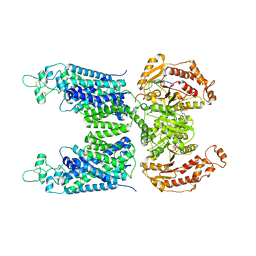 | | Cryo-EM structure of human NCC (class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polythiazide, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHO
 
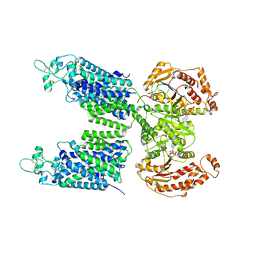 | | Cryo-EM structure of human NCC (class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polythiazide, SODIUM ION, ... | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHR
 
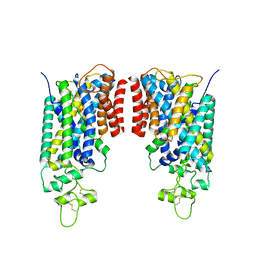 | | Cryo-EM structure of human NCC (class 3-3) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHP
 
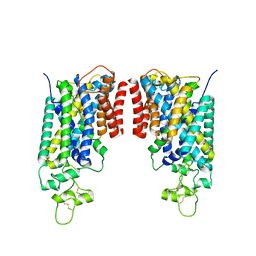 | | Cryo-EM structure of human NCC (class 3-1) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHT
 
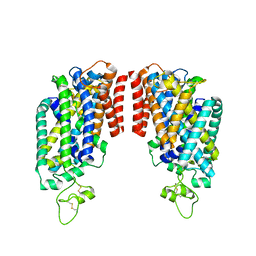 | | Cryo-EM structure of human NCC | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 12 member 3 | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
8FHQ
 
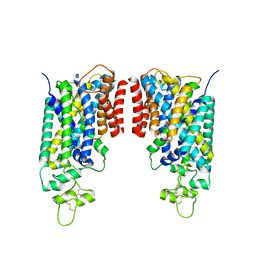 | | Cryo-EM structure of human NCC (class 3-2) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
7NSL
 
 | |
6ZCG
 
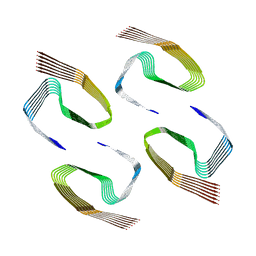 | |
6Z1I
 
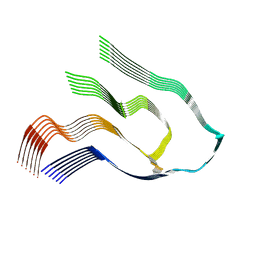 | |
6Z1O
 
 | |
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
6FAN
 
 | |
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
6SHS
 
 | | Abeta fibril (Morphology I) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Kollmer, M, Fandrich, M. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure and polymorphism of A beta amyloid fibrils purified from Alzheimer's brain tissue.
Nat Commun, 10, 2019
|
|
