6RA6
 
 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
7OHD
 
 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
8PF4
 
 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)(4-fluorophenyl)carbamoyl)-1-methyl-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Exertier, C, Ilari, A, Fiorillo, A, Antonelli, L. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
8PF3
 
 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 1-(3,4-dichlorobenzyl)-4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)(4-fluorophenyl)carbamoyl)-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | 4-[(3,4-dichlorophenyl)methyl]-~{N}-(4-fluorophenyl)-~{N}-[[5-[2-(4-fluorophenyl)ethylcarbamoyl]furan-2-yl]methyl]-4-(3-phenylpropyl)-1,4$l^{4}-diazinane-1-carboxamide, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Exertier, C, Ilari, A, Fiorillo, A, Antonelli, L. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
8PF5
 
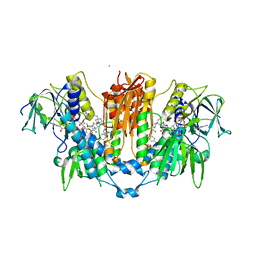 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 1-(3,4-dichlorobenzyl)-4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)carbamoyl)-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | 4-[(3,4-dichlorophenyl)methyl]-~{N}-[[5-[2-(4-fluorophenyl)ethylcarbamoyl]furan-2-yl]methyl]-4-(3-phenylpropyl)-1,4$l^{4}-diazinane-1-carboxamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Exertier, C, Antonelli, L, Fiorillo, A, Ilari, A. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
6H6J
 
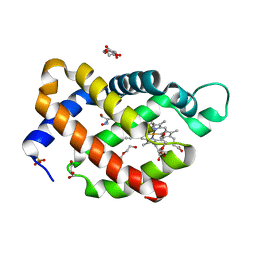 | | Carbomonoxy murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBON MONOXIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H5Z
 
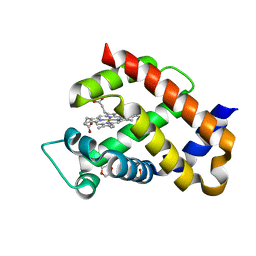 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6I
 
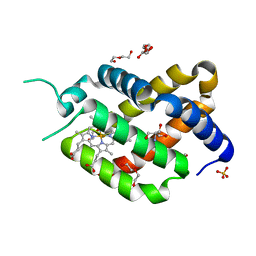 | | Ferric murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6C
 
 | | Carbomonoxy murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
5NHT
 
 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
5NQK
 
 | | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6I3T
 
 | | Crystal structure of murine neuroglobin bound to CO at 40 K. | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C. | | Deposit date: | 2018-11-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
7Q6X
 
 | | OleP mutant S240Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q89
 
 | | OleP mutant G92W in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q6R
 
 | | OleP mutant E89Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7ZTH
 
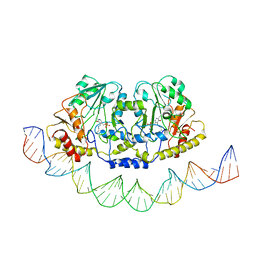 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
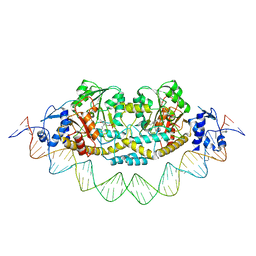 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
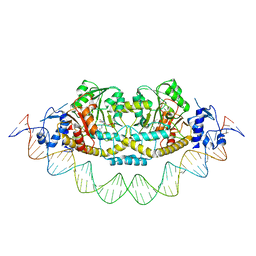 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
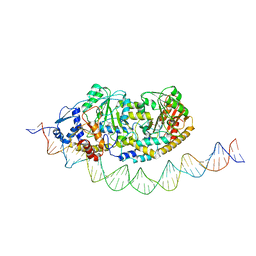 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
