2I9T
 
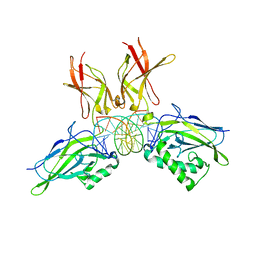 | | Structure of NF-kB p65-p50 heterodimer bound to PRDII element of B-interferon promoter | | Descriptor: | 5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*CP*TP*G)-3', 5'-D(*CP*AP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*CP*T)-3', Nuclear factor NF-kappa-B p105 subunit, ... | | Authors: | Escalante, C.R, Shen, L, Thanos, D, Aggarwal, A.K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of NF-kappaB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-beta promoter
Structure, 10, 2002
|
|
7JSG
 
 | | Adeno-Associated Virus 2 Rep68 HD-Heptamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSH
 
 | | Adeno-Associated Virus 2 Rep68 HD Heptamer-ssAAVS1 with ATPgS | | Descriptor: | DNA (5'-D(P*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*C)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSE
 
 | | Adeno-Associated Virus Origin Binding Domain in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSF
 
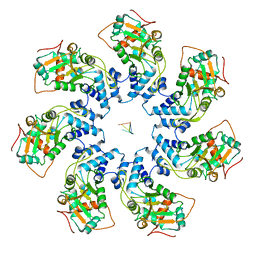 | | Adeno-Associated Virus Helicase domain Heptamer with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSI
 
 | | Adeno-Associated Virus 2 Rep68 HD Hexamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
1IF1
 
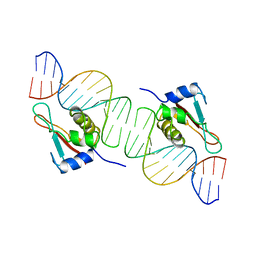 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
2PI0
 
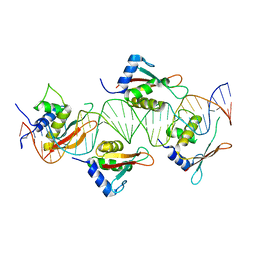 | | Crystal Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-B enhancer | | Descriptor: | Interferon regulatory factor 3, PRDIII-I region of human interferon-B promoter strand 1, PRDIII-I region of human interferon-B promoter strand 2 | | Authors: | Escalante, C.R, Nistal-Villan, E, Leyi, S, Garcia-Sastre, A, Aggarwal, A.K. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer.
Mol.Cell, 26, 2007
|
|
6XB8
 
 | |
1U0J
 
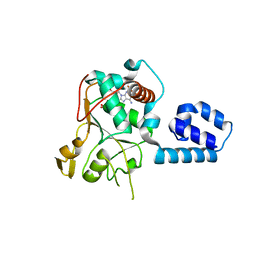 | | Crystal Structure of AAV2 Rep40-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein | | Authors: | James, J.A, Aggarwal, A.K, Linden, R.M, Escalante, C.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of adeno-associated virus type 2 Rep40-ADP complex: Insight into
nucleotide recognition and catalysis by superfamily 3 helicases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5EYB
 
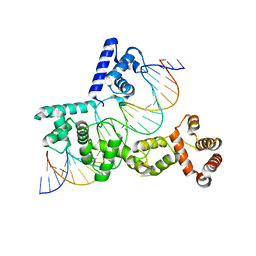 | | X-ray Structure of Reb1-Ter Complex | | Descriptor: | DNA (26-MER), DNA-binding protein reb1 | | Authors: | Jaiswal, R, Choudhury, M, Zaman, S, Singh, S, Santosh, V, Bastia, D, Escalante, C.R. | | Deposit date: | 2015-11-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BYG
 
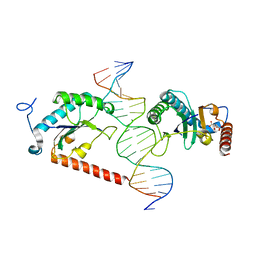 | | X-ray structure of AAV2 OBD-AAVS1 complex 2:1 | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Musayev, F.N, Escalante, C.R. | | Deposit date: | 2015-06-10 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Assembly of the Adeno-associated Virus Type 2 Rep68 Protein on the Integration Site AAVS1.
J.Biol.Chem., 290, 2015
|
|
6NP6
 
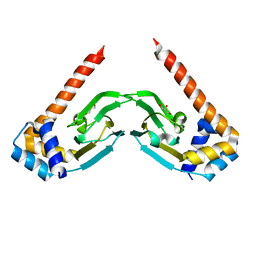 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5V30
 
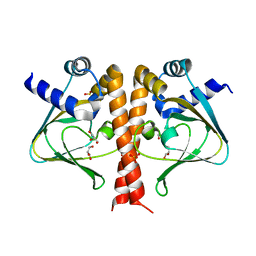 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4ZQ9
 
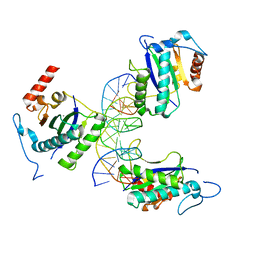 | | X-ray structure of AAV-2 OBD bound to AAVS1 site 3:1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*CP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*GP*GP*GP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Musayev, F.N, Zarate-Perez, F, Escalante, C.R. | | Deposit date: | 2015-05-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Assembly of the Adeno-associated Virus Type 2 Rep68 Protein on the Integration Site AAVS1.
J.Biol.Chem., 290, 2015
|
|
4ZO0
 
 | |
3QU6
 
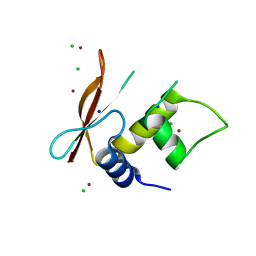 | | Crystal structure of IRF-3 DBD free form | | Descriptor: | CHLORIDE ION, IRF3 protein, SODIUM ION, ... | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3QU3
 
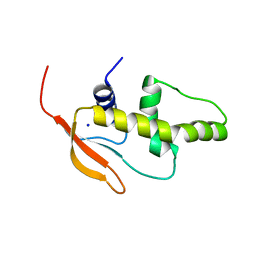 | | Crystal structure of IRF-7 DBD apo form | | Descriptor: | 1,2-ETHANEDIOL, Interferon regulatory factor 7, SODIUM ION | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
1S97
 
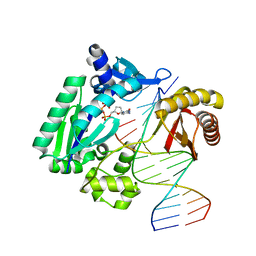 | | DPO4 with GT mismatch | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*G)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SDO
 
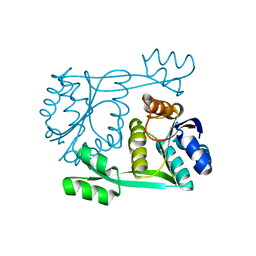 | | Crystal Structure of Restriction Endonuclease BstYI | | Descriptor: | BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Vanamee, E.S, Edwards, T.A, Escalante, C.R, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2004-02-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of BstYI at 1.85 A Resolution: A Thermophilic Restriction Endonuclease with Overlapping Specificities to BamHI and BglII
J.Mol.Biol., 338, 2004
|
|
1S9F
 
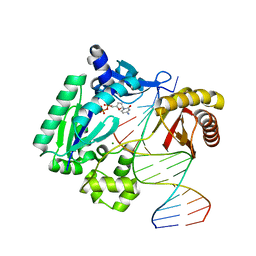 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S9H
 
 | | Crystal Structure of Adeno-associated virus Type 2 Rep40 | | Descriptor: | Rep 40 protein | | Authors: | James, J.A, Escalante, C.R, Yoon-Robarts, M, Edwards, T.A, Linden, R.M, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the SF3 Helicase from Adeno-Associated Virus Type 2
Structure, 11, 2003
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
1JIH
 
 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
