3MEF
 
 | |
6F0G
 
 | | Crystal structure ASF1-ip3 | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, ip3 | | Authors: | Gaubert, A, Guichard, B, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
6F0H
 
 | | Crystal structure ASF1-ip4 | | Descriptor: | CITRIC ACID, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
7KVC
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
3O74
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
7R4Q
 
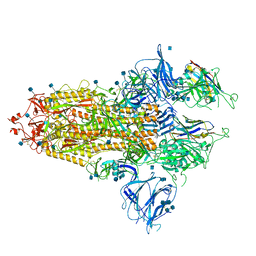 | | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.29, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4I
 
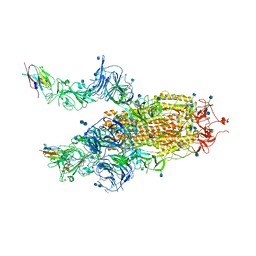 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4R
 
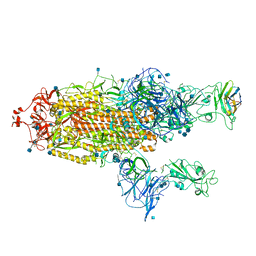 | | The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.10, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
2UY5
 
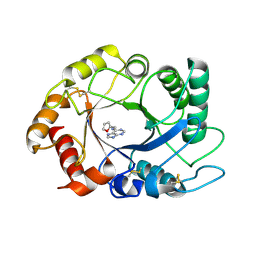 | | ScCTS1_kinetin crystal structure | | Descriptor: | ENDOCHITINASE, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2UY2
 
 | | ScCTS1_apo crystal structure | | Descriptor: | ENDOCHITINASE, GLYCEROL | | Authors: | Hurtado-Guerrero, R, Van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2UY4
 
 | | ScCTS1_acetazolamide crystal structure | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ENDOCHITINASE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2VXK
 
 | | Structural comparison between Aspergillus fumigatus and human GNA1 | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE 6-PHOSPHATE ACETYLTRANSFERASE, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O.G, Min, J, Zeng, H, Vallius, L, Shepherd, S, Ibrahim, A.F.M, Wu, H, Plotnikov, A.N, van Aalten, D.M.F. | | Deposit date: | 2008-07-05 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Differences between Human and Aspergillus Fumigatus D-Glucosamine-6- Phosphate N-Acetyltransferase.
Biochem.J., 415, 2008
|
|
6GW7
 
 | | The CTD of HpDprA, a DNA binding Winged Helix domain which do not bind dsDNA | | Descriptor: | DNA protecting protein DprA | | Authors: | Lisboa, J, Celma, L, Sanchez, D, Marquis, M, Andreani, J, Guerois, R, Ochsenbein, F, Durand, D, Marsin, S, Cuniasse, P, Radicella, J.P, Quevillon-Cheruel, S. | | Deposit date: | 2018-06-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of HpDprA is a DNA-binding winged helix domain that does not bind double-stranded DNA.
Febs J., 286, 2019
|
|
2VYO
 
 | | Chitin deacetylase family member from Encephalitozoon cuniculi | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Urch, J.E, Hurtado-Guerrero, R, Texier, C, Van Aalten, D.M.F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Putative Polysaccharide Deacetylase of the Human Parasite Encephalitozoon Cuniculi.
Protein Sci., 18, 2009
|
|
2UY3
 
 | |
2W63
 
 | |
2W61
 
 | |
2W62
 
 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
2VEZ
 
 | | AfGNA1 crystal structure complexed with Acetyl-CoA and Glucose-6P gives new insights into catalysis | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, PHOSPHATE ION, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O, Shepherd, S, van Aalten, D.M.F. | | Deposit date: | 2007-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Glucose-6-Phosphate as a Probe for the Glucosamine- 6-Phosphate N-Acetyltransferase Michaelis Complex.
FEBS Lett., 581, 2007
|
|
2WZG
 
 | | Legionella glucosyltransferase (Lgt1) crystal structure | | Descriptor: | GLUCOSYLTRANSFERASE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Hurtado-Guerrero, R, Zusman, T, Pathak, S, Ibrahim, A.F.M, Shepherd, S, Prescott, A, Segal, G, Van Aalten, D.M.F. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of Elongation Factor 1A Inhibition by a Legionella Pneumophila Glycosyltransferase.
Biochem.J., 426, 2010
|
|
2WZF
 
 | | Legionella pneumophila glucosyltransferase crystal structure | | Descriptor: | GLUCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Hurtado-Guerrero, R, Zusman, T, Pathak, S, Ibrahim, A.F.M, Shepherd, S, Prescott, A, Segal, G, van Aalten, D.M.F. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Elongation Factor 1A Inhibition by a Legionella Pneumophila Glycosyltransferase.
Biochem.J., 426, 2010
|
|
4B4D
 
 | | Crystal structure of FAD-containing ferredoxin-NADP reductase from Xanthomonas axonopodis pv. citri | | Descriptor: | CHLORIDE ION, FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Martinez-Julvez, M, Orellano, E.G, Tondo, M.L, Hurtado-Guerrero, R, Medina, M, Ceccarelli, E.A, Sanchez-Azqueta, A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Fad-Containing Ferredoxin- Nadp Reductase from Xanthomonas Axonopodis Pv. Citri
Biomed Res Int, 2013, 2013
|
|
3GTZ
 
 | | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium | | Descriptor: | GLYCEROL, Putative translation initiation inhibitor | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-28 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium
To be Published
|
|
