6PEL
 
 | |
6PGS
 
 | |
6PH7
 
 | | Crystal structure of bovine opsin with nerol bound | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dien-1-ol, G protein CT2 peptide, PALMITIC ACID, ... | | Authors: | Eger, B.T, Morizumi, T, Ernst, O.P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Odorant-binding site in visual opsin
To Be Published
|
|
6NWE
 
 | |
5ITE
 
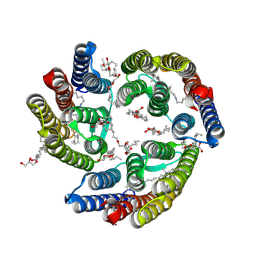 | | 2.2-Angstrom in meso crystal structure of Haloquadratum Walsbyi Bacteriorhodopsin (HwBR) from Octylglucoside (OG) Detergent Micelles | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | Authors: | Broecker, J, Eger, B.T, Ernst, O.P. | | Deposit date: | 2016-03-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs.
Structure, 25, 2017
|
|
5ITC
 
 | | 2.2-Angstrom in meso crystal structure of Haloquadratum Walsbyi Bacteriorhodopsin (HwBR) from Styrene Maleic Acid (SMA) Polymer Nanodiscs | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | Authors: | Broecker, J, Eger, B.T, Ernst, O.P. | | Deposit date: | 2016-03-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs.
Structure, 25, 2017
|
|
5JGN
 
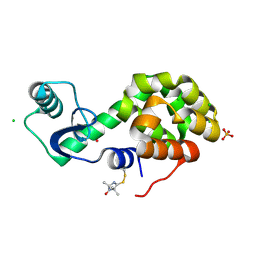 | | Spin-Labeled T4 Lysozyme Construct I9V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JGR
 
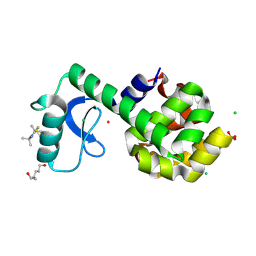 | | Spin-Labeled T4 Lysozyme Construct K43V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JGX
 
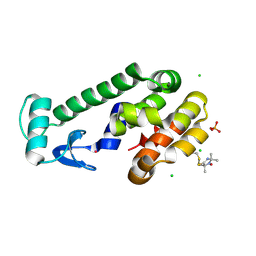 | | Spin-Labeled T4 Lysozyme Construct V131V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JOM
 
 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5KGR
 
 | | Spin-Labeled T4 Lysozyme Construct I9V1/V131V1 (30 days) | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-06-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JGZ
 
 | | Spin-Labeled T4 Lysozyme Construct T151V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5KKI
 
 | | 1.7-Angstrom in situ Mylar structure of hen egg-white lysozyme (HEWL) at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5JGU
 
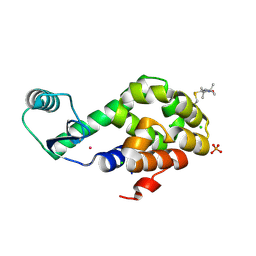 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5KKJ
 
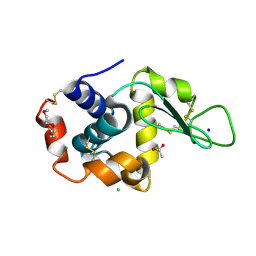 | | 2.0-Angstrom In situ Mylar structure of hen egg-white lysozyme (HEWL) at 293 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5JGV
 
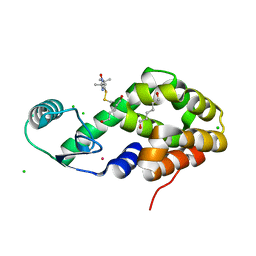 | | Spin-Labeled T4 Lysozyme Construct A73V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5KKH
 
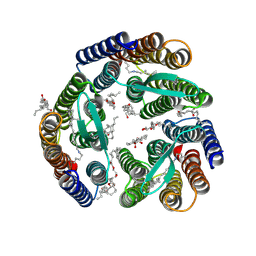 | | 2.1-Angstrom In situ Mylar structure of bacteriorhodopsin from Haloquadratum walsbyi (HwBR) at 100 K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5KKK
 
 | |
