8Q54
 
 | |
8Q3V
 
 | | Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex | | Descriptor: | MAGNESIUM ION, SODIUM ION, Tetrahydromethanopterin S-methyltransferase subunit A 1, ... | | Authors: | Aziz, I, Vonck, J, Ermler, U. | | Deposit date: | 2023-08-04 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural and mechanistic basis of the central energy-converting methyltransferase complex of methanogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5T5M
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRIGONAL FORM AT 2.5 A. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5T5I
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, ORTHORHOMBIC FORM AT 1.9 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
4DPM
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota in complex with CoA | | Descriptor: | COENZYME A, MAGNESIUM ION, Malonyl-CoA/succinyl-CoA reductase | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
6QKX
 
 | |
6QKR
 
 | |
6QKG
 
 | | 2-Naphthoyl-CoA Reductase(NCR) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kayastha, K, Ermler, U. | | Deposit date: | 2019-01-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low potential enzymatic hydride transfer via highly cooperative and inversely functionalized flavin cofactors.
Nat Commun, 10, 2019
|
|
4DPK
 
 | | Structure of malonyl-coenzyme A reductase from crenarchaeota | | Descriptor: | Malonyl-CoA/succinyl-CoA reductase, PHOSPHATE ION | | Authors: | Demmer, U, Warkentin, E, Srivastava, A, Kockelkorn, D, Fuchs, G, Ermler, U. | | Deposit date: | 2012-02-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for a Bispecific NADP+ and CoA Binding Site in an Archaeal Malonyl-Coenzyme A Reductase.
J.Biol.Chem., 288, 2013
|
|
2I2X
 
 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3F47
 
 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
2FHK
 
 | | Crystal structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, N-[4,5,7-TRICARBOXYHEPTANOYL]-L-GAMMA-GLUTAMYL-N-{2-[4-({5-[(FORMYLAMINO)METHYL]-3-FURYL}METHOXY)PHENYL]ETHYL}-D-GLUTAMINE, POTASSIUM ION | | Authors: | Acharya, P, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2005-12-25 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of formylmethanofuran: tetrahydromethanopterin formyltransferase in complex with its coenzymes
J.Mol.Biol., 357, 2006
|
|
5HLR
 
 | | Linalool dehydratase/isomerase: Ldi-apo | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Linalool dehydratase/isomerase | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
5HSS
 
 | | Linalool dehydratase/isomerase: Ldi with monoterpene substrate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-Myrcene, Geraniol, ... | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-26 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
5L8X
 
 | |
5LAA
 
 | |
1JNR
 
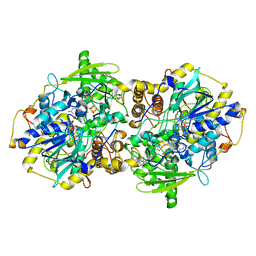 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2001-07-25 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1Y5Y
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 | | Descriptor: | CALCIUM ION, Formaldehyde-activating enzyme fae, SODIUM ION | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1Z47
 
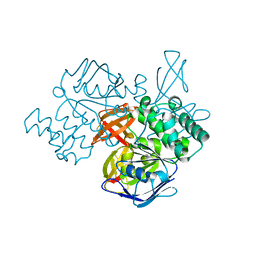 | | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | | Descriptor: | CHLORIDE ION, putative ABC-transporter ATP-binding protein | | Authors: | Scheffel, F, Demmer, U, Warkentin, E, Huelsmann, A, Schneider, E, Ermler, U. | | Deposit date: | 2005-03-15 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Febs Lett., 579, 2005
|
|
1JNZ
 
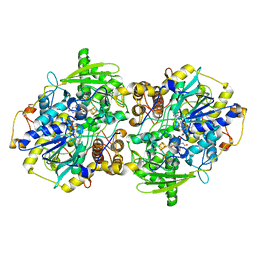 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, SULFITE ION, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M, Ermler, U. | | Deposit date: | 2001-07-26 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8QPL
 
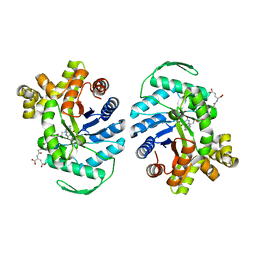 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QPM
 
 | | Structure of methylene-tetrahydromethanopterin reductase from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QPJ
 
 | |
8QQ8
 
 | | Crystal Structure of F420-dependent Methylene-Tetrahydromethanopterin Reductase Mutant E6Q from Methanocaldococcus Jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
