4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
7BKW
 
 | | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound
To Be Published
|
|
7BKV
 
 | | Endothiapepsin structure obtained at 100K with fragment AC39729 bound | | Descriptor: | 5-fluoranylpyridin-2-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment AC39729 bound
To Be Published
|
|
7BKS
 
 | | 100K endothiapepsin structure obtained in presence of 40 mM DMSO | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | 100K endothiapepsin structure obtained in presence of 40 mM DMSO
To Be Published
|
|
7BKU
 
 | | Endothiapepsin structure obtained at 100K with fragment JFD03909 bound | | Descriptor: | 1,10-PHENANTHROLINE, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment JFD03909 bound
To Be Published
|
|
7NT2
 
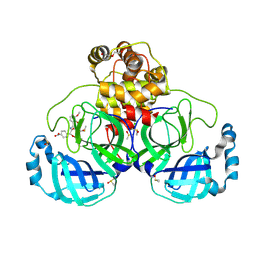 | | Crystal structure of SARS CoV2 main protease in complex with FSP006 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT1
 
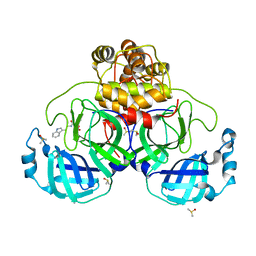 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT3
 
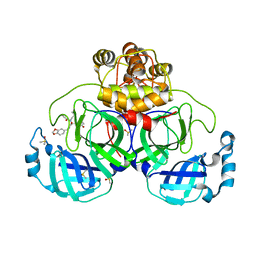 | | Crystal structure of SARS CoV2 main protease in complex with FSCU015 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTV
 
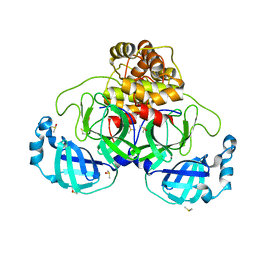 | | Crystal structure of SARS CoV2 main protease in complex with DN_EG_002 (modelled using PanDDA event map) | | Descriptor: | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NUK
 
 | | Crystal structure of SARS CoV2 main protease in complex with EG009 (modelled using PanDDA event map) | | Descriptor: | 2-[2-chloranylethanoyl(propyl)amino]-~{N}-(2-methoxyphenyl)ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6G2R
 
 | | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]-2,3,5,6-tetrakis(fluoranyl)benzenecarbonitrile, SULFATE ION, ... | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
2ETN
 
 | | Crystal structure of Thermus aquaticus Gfh1 | | Descriptor: | anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Lamour, V, Hogan, B.P, Erie, D.A, Darst, S.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage.
J.Mol.Biol., 356, 2006
|
|
