1TGV
 
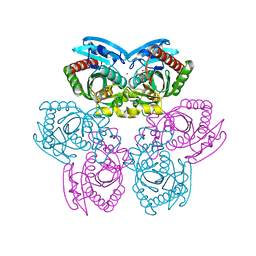 | | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate | | Descriptor: | 5-FLUOROURIDINE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Bu, W, Settembre, E.C, Sanders, J.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of E. coli Uridine Phosphorylase
To be Published, 2004
|
|
1R9G
 
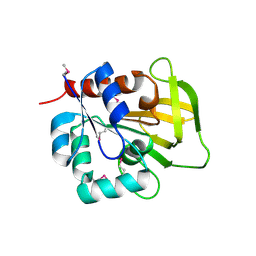 | | Three-dimensional Structure of YaaE from Bacillus subtilis | | Descriptor: | Hypothetical protein yaaE | | Authors: | Bauer, J.A, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-10-29 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of YaaE from Bacillus subtilis, a Glutaminase Implicated in Pyridoxal-5'-phosphate Biosynthesis.
J.Biol.Chem., 279, 2004
|
|
1TWJ
 
 | |
1TLU
 
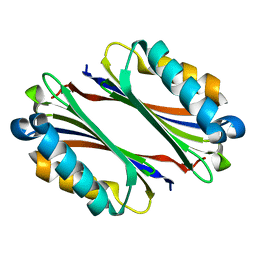 | | Crystal Structure of Thermotoga maritima S-adenosylmethionine decarboxylase | | Descriptor: | S-adenosylmethionine decarboxylase proenzyme, AdoMetDC, SamDC | | Authors: | Toms, A.V, Kinsland, C, McCloskey, D.E, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evolutionary Links as Revealed by the Structure of Thermotoga maritima S-Adenosylmethionine Decarboxylase.
J.Biol.Chem., 279, 2004
|
|
1TMI
 
 | | Structure of Thermotoga maritima S63A non-processing mutant S-adenosylmethionine decarboxylase | | Descriptor: | S-adenosylmethionine decarboxylase proenzyme, AdoMetDC, SamDC | | Authors: | Toms, A.V, Kinsland, C, McCloskey, D.E, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary Links as Revealed by the Structure of Thermotoga maritima S-Adenosylmethionine Decarboxylase.
J.Biol.Chem., 279, 2004
|
|
1U1F
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(m-(benzyloxy)benzyl)acyclouridine (BBAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(3-(BENZYLOXY)BENZYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U11
 
 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TPT
 
 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
2ATE
 
 | | Structure of the complex of PurE with NitroAIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kappock, T.J, Mathews, I.I, Zaugg, J.B, Peng, P, Hoskins, A.A, Okamoto, A, Ealick, S.E, Stubbe, J. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
Biochemistry, 46, 2007
|
|
2SN3
 
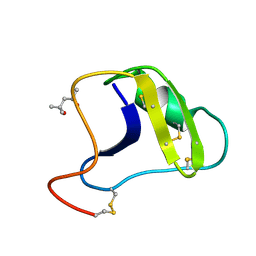 | | STRUCTURE OF SCORPION TOXIN VARIANT-3 AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SCORPION NEUROTOXIN (VARIANT 3) | | Authors: | Zhao, B, Carson, M, Ealick, S.E, Bugg, C.E. | | Deposit date: | 1992-02-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of scorpion toxin variant-3 at 1.2 A resolution.
J.Mol.Biol., 227, 1992
|
|
2TPT
 
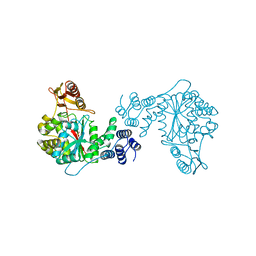 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
2THI
 
 | |
3C9S
 
 | | AaThiL complexed with AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Thiamine monophosphate kinase | | Authors: | McCulloch, K.M, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of thiamin monophosphate kinase in complex with substrates and products.
Biochemistry, 47, 2008
|
|
3C9T
 
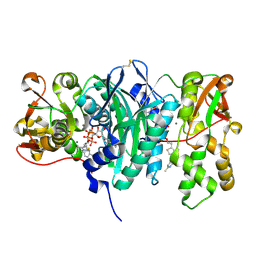 | | AaThiL complexed with AMPPCP and TMP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, THIAMIN PHOSPHATE, ... | | Authors: | McCulloch, K.M, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of thiamin monophosphate kinase in complex with substrates and products.
Biochemistry, 47, 2008
|
|
3C9U
 
 | | AaThiL complexed with ADP and TPP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | McCulloch, K.M, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural studies of thiamin monophosphate kinase in complex with substrates and products.
Biochemistry, 47, 2008
|
|
2AI3
 
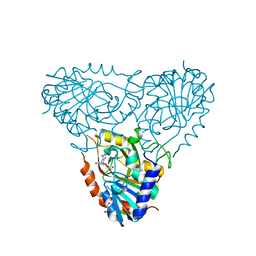 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | (2S,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YLPHOSPHONI C ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AI1
 
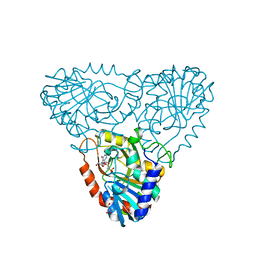 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | ((2R,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YL)METHYLPHOSPHONIC ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2QCX
 
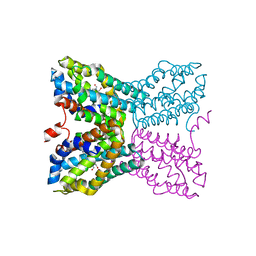 | | Crystal structure of Bacillus subtilis TenA Y112F mutant complexed with formyl aminomethyl pyrimidine | | Descriptor: | N-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-N-(2-HYDROXYETHYL)FORMAMIDE, Transcriptional activator tenA | | Authors: | Zhang, Y, Jenkins, A.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Bioorg.Chem., 36, 2008
|
|
2R7M
 
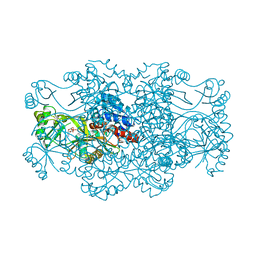 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMP | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R84
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP and AICAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7N
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | Descriptor: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R87
 
 | |
2TPS
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PROTEIN (THIAMIN PHOSPHATE SYNTHASE), PYROPHOSPHATE 2-, ... | | Authors: | Chiu, H.-J, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
Biochemistry, 38, 1999
|
|
2R85
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
